|
|
--- |
|
|
license: apache-2.0 |
|
|
tags: |
|
|
- biology |
|
|
- chemistry |
|
|
- biomolecular-structure-prediction |
|
|
- IntelliFold |
|
|
--- |
|
|
|
|
|
 |
|
|
|
|
|
# IntelliFold: A Controllable Foundation Model for General and Specialized Biomolecular Structure Prediction. |
|
|
[](https://huggingface.co/GAGABIG/CNN) |
|
|
[](https://pypi.org/project/intellifold/) |
|
|
[](LICENSE) |
|
|
[](#contact-us) |
|
|
|
|
|
|
|
|
<div align="center" style="margin: 20px 0;"> |
|
|
<span style="margin: 0 10px;">⚡ <a href="https://server.intfold.com">IntelliFold Server</a></span> |
|
|
• <span style="margin: 0 10px;">📄 <a href="https://arxiv.org/abs/2507.02025">Technical Report</a></span> |
|
|
</div> |
|
|
|
|
|
|
|
|
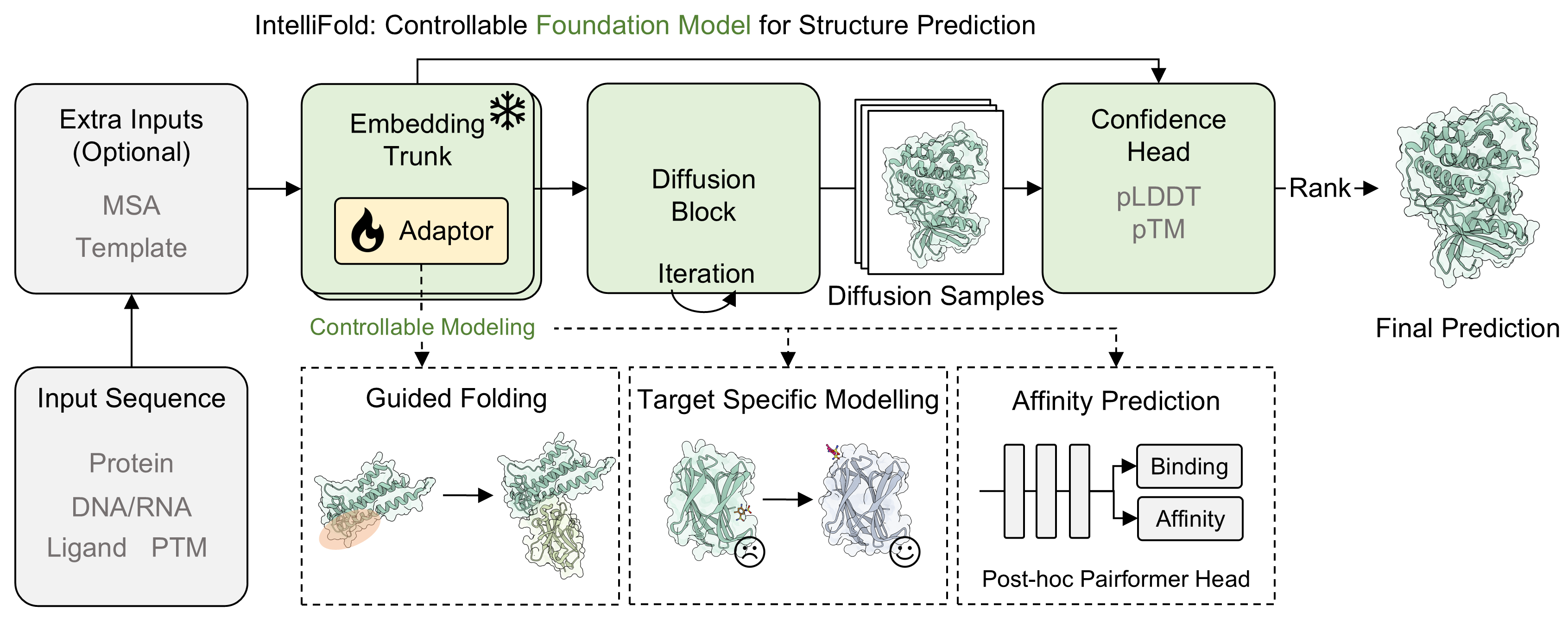 |
|
|
|
|
|
|
|
|
## 🚀 Quick Start |
|
|
|
|
|
To quickly get started with IntelliFold, you can use the following commands: |
|
|
```bash |
|
|
# Install IntelliFold from PyPI |
|
|
pip install intellifold |
|
|
# Run inference with an example YAML file |
|
|
intellifold predict ./examples/5S8I_A.yaml --out_dir ./output |
|
|
``` |
|
|
|
|
|
## ⚙️ Installation |
|
|
|
|
|
To more complete installation instructions and usage, please refer to the [Installation Guide](https://github.com/IntelliGen-AI/IntelliFold/blob/main/docs/installation.md). |
|
|
|
|
|
|
|
|
## 🔍 Inference |
|
|
|
|
|
1. **Prepare Input File**: Create a YAML file with your sequences following our [input format specification](https://github.com/IntelliGen-AI/IntelliFold/blob/main/docs/input_yaml_format.md) |
|
|
|
|
|
2. **Run Prediction**: |
|
|
```bash |
|
|
intellifold predict your_input.yaml --out_dir ./results |
|
|
``` |
|
|
|
|
|
3. **Check Results**: Find predicted structures and confidence scores in the output directory, you can also check the section of **output format** in [output documentation](https://github.com/IntelliGen-AI/IntelliFold/blob/main/docs/input_yaml_format.md#output-format). |
|
|
|
|
|
4. **Optional Optimization**: Enable [custom kernels](https://github.com/IntelliGen-AI/IntelliFold/blob/main/docs/kernels.md) for faster inference and reduced memory usage |
|
|
|
|
|
For comprehensive usage instructions and examples, refer to the [Usage Guide](https://github.com/IntelliGen-AI/IntelliFold/blob/main/docs/usage.md). |
|
|
|
|
|
|
|
|
## 📊 Benchmarking |
|
|
To comprehensively evaluate the performance of To quickly get started with IntelliFold, you can use the following commands: |
|
|
, we conducted a rigorous evaluation on [FoldBench](https://github.com/BEAM-Labs/FoldBench). We compared IntelliFold against several leading methods, including [Boltz-1,2](https://github.com/jwohlwend/boltz), [Chai-1](https://github.com/chaidiscovery/chai-lab), [Protenix](https://github.com/bytedance/Protenix) and [Alphafold3](https://github.com/google-deepmind/alphafold3). |
|
|
|
|
|
For more details on the benchmarking process and results, please refer to our [Technical Report](https://arxiv.org/abs/2507.02025). |
|
|
|
|
|
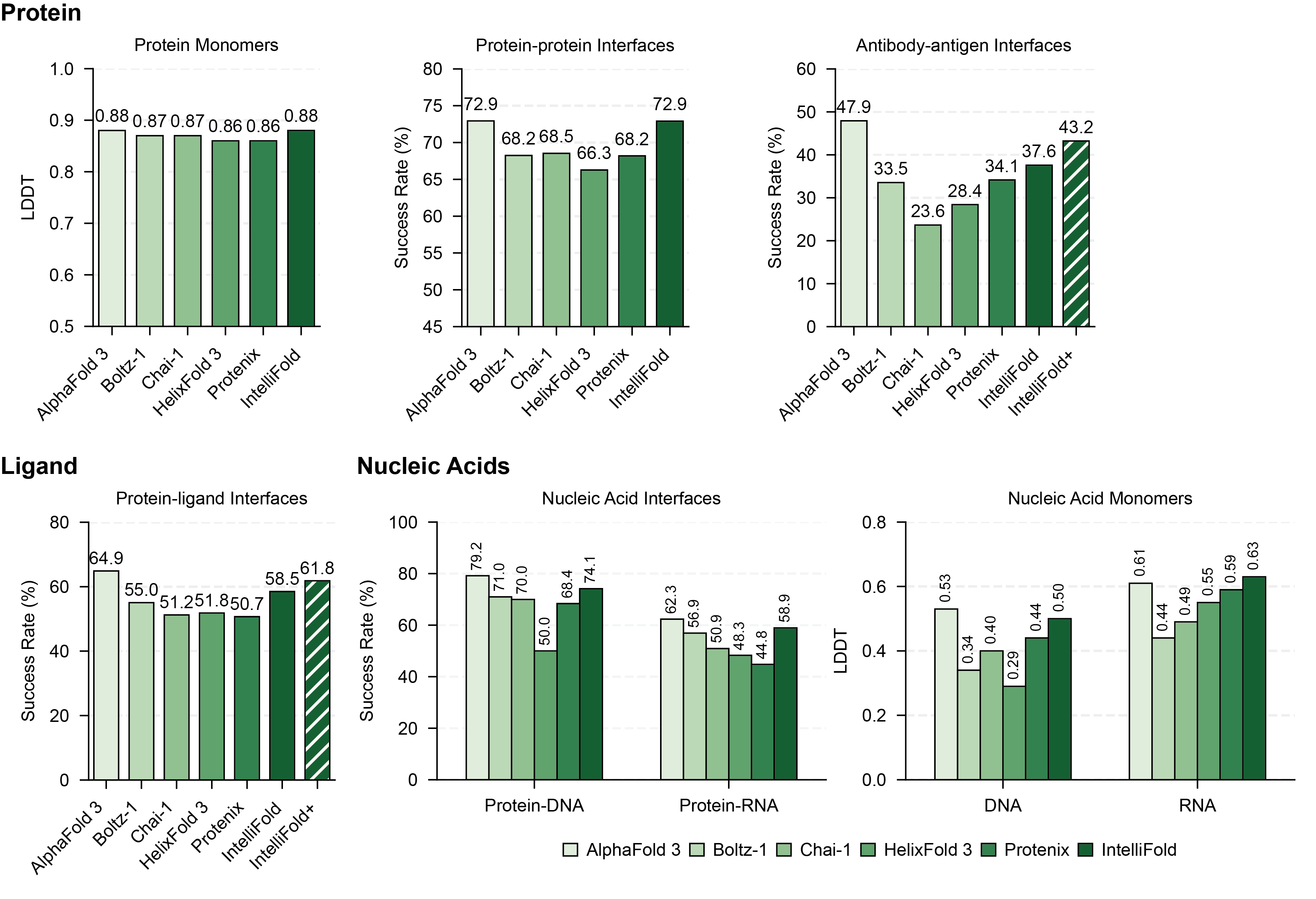 |
|
|
|
|
|
|
|
|
## 🌐 IntelliFold Server |
|
|
|
|
|
**We highly recommend using the [IntelliFold Server](https://server.intfold.com) for the most accurate, complete, and convenient biomolecular structure predictions.** It requires no installation and provides an intuitive web interface to submit your sequences and visualize results directly in your browser. The server runs the **full, optimized, latest** IntelliFold implementation for optimal performance. |
|
|
|
|
|
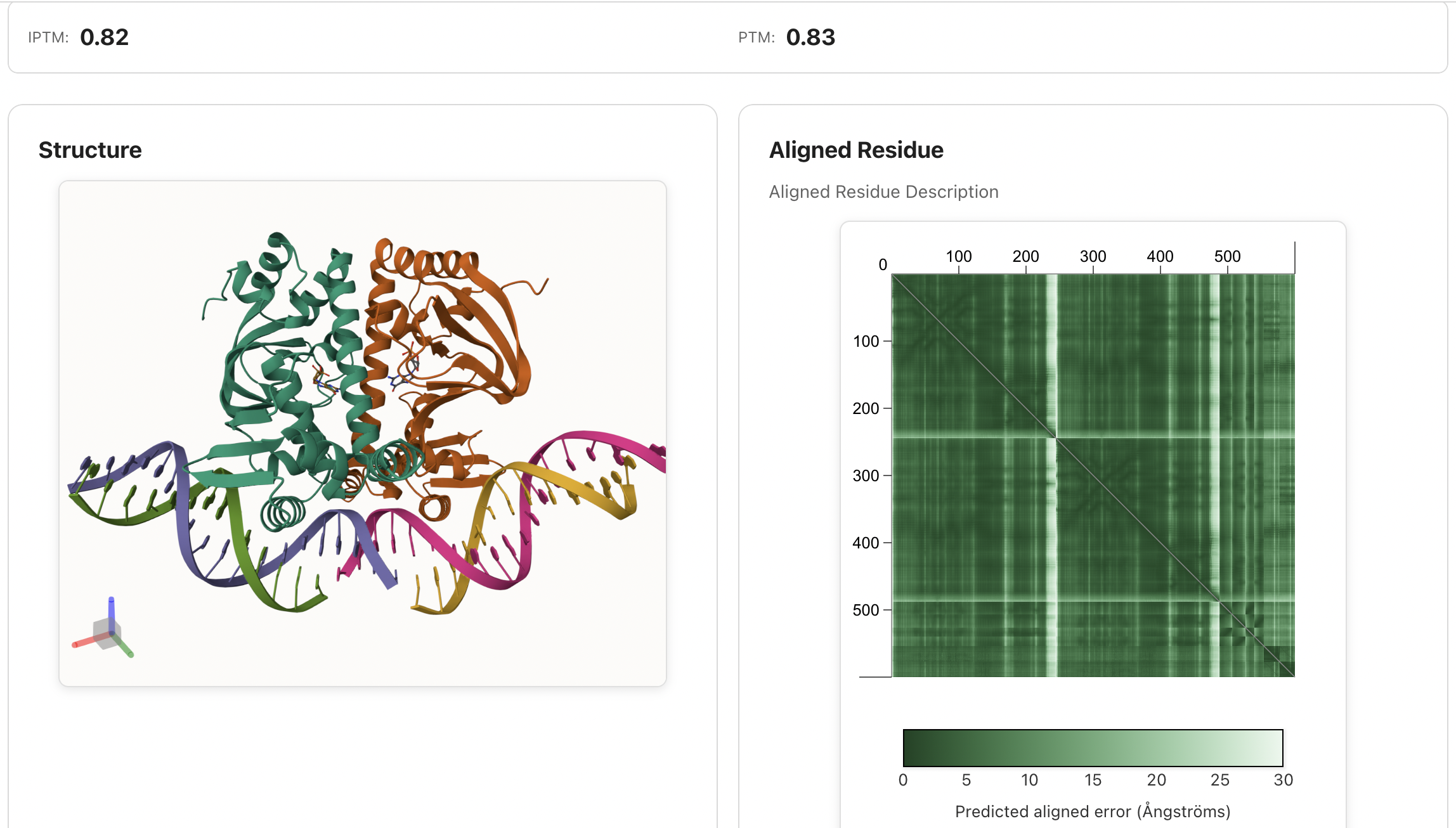 |
|
|
|
|
|
|
|
|
## 📜 Citation |
|
|
|
|
|
If you use IntelliFold in your research, please cite our paper: |
|
|
|
|
|
``` |
|
|
@misc{theintfoldteam2025intfoldcontrollablefoundationmodel, |
|
|
title={IntFold: A Controllable Foundation Model for General and Specialized Biomolecular Structure Prediction}, |
|
|
author={The IntFold Team and Leon Qiao and Wayne Bai and He Yan and Gary Liu and Nova Xi and Xiang Zhang}, |
|
|
year={2025}, |
|
|
eprint={2507.02025}, |
|
|
archivePrefix={arXiv}, |
|
|
primaryClass={q-bio.BM}, |
|
|
url={https://arxiv.org/abs/2507.02025} |
|
|
} |
|
|
``` |
|
|
|
|
|
## 🔗 Acknowledgements |
|
|
|
|
|
- The implementation of **fast layernorm operators** is inspired by [OneFlow](https://github.com/Oneflow-Inc/oneflow) and [FastFold](https://github.com/hpcaitech/FastFold), following [Protenix](https://github.com/bytedance/Protenix)'s usage. |
|
|
- Many components in `intellifold/openfold/` are adapted from [OpenFold](https://github.com/aqlaboratory/openfold), with substantial modifications and improvements by our team (except for the `LayerNorm` part). |
|
|
- This repository, the implementation of **Inference Data Pipeline**(Data/Feature Processing and MSA generation tasks) referred to [Boltz-1](https://github.com/jwohlwend/boltz), and modify some codes to adapt to the input of our model. |
|
|
|
|
|
|
|
|
|
|
|
## ⚖️ License |
|
|
|
|
|
The IntelliFold project, including code and model parameters, is made available under the [Apache 2.0 License](https://github.com/IntelliGen-AI/IntelliFold/blob/main/LICENSE), it is free for both academic research and commercial use. |
|
|
|
|
|
## 📬 Contact Us |
|
|
|
|
|
If you have any questions or are interested in collaboration, please feel free to contact us at [email protected]. |