APOLO Medical Multimodal Instruct
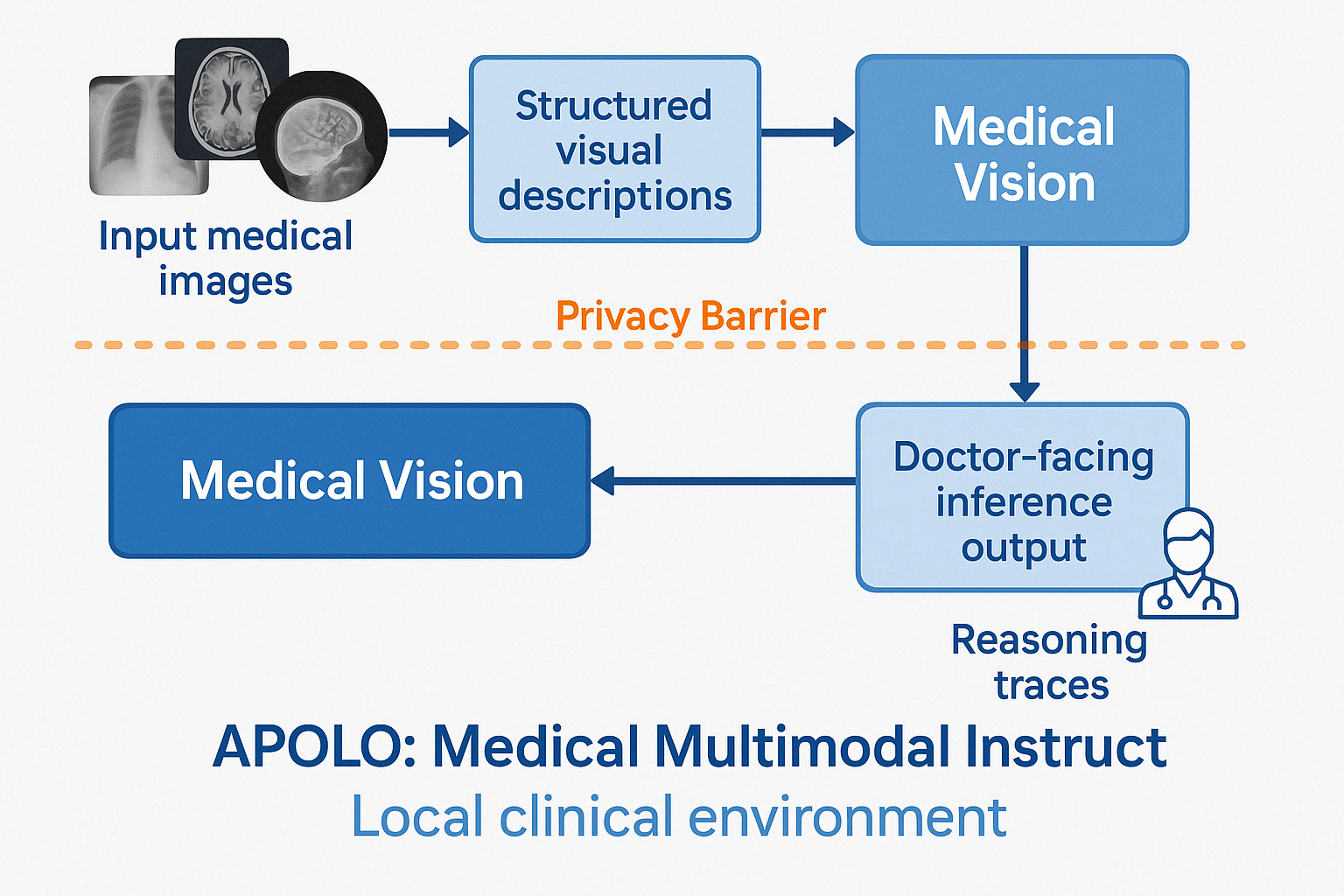
APOLO Medical Multimodal Instruct is a privacy-preserving multimodal model combining vision and language capabilities to process medical images while maintaining strict data protection. Built upon the DeepSeek-VL2-tiny architecture, this model implements a novel two-stage processing pipeline that ensures patient privacy while enabling advanced diagnostic reasoning.
Model Details
Model Description
APOLO Medical Multimodal Instruct is designed specifically for clinical environments requiring both robust medical image interpretation and strong privacy guarantees. The model combines an asynchronous visual description pipeline with a diagnostic reasoning engine in a unified architecture while maintaining logical separation between these components.
- Developed by: APOLO AI Research Team
- Model type: Multimodal Vision-Language Model (based on DeepSeek-VL2-tiny)
- Language(s): English, Chinese
- License: Apache 2.0 with additional healthcare compliance provisions
- Finetuned from model: DeepSeek-VL2-tiny (1.0B activated parameters)
Model Architecture
APOLO Medical Multimodal Instruct implements a unique two-stage architecture within a single model:
Stage 1: APOLO Medical Vision
- Processes medical images to generate structured visual descriptions
- Operates asynchronously and continuously
- Uses dynamic tiling strategy for high-resolution medical images
- Designed to capture clinically relevant visual information without patient identifiers
Stage 2: APOLO Medical Instruct
- Performs diagnostic reasoning based only on the structured descriptions
- Operates on-demand when clinicians request insights
- Includes explainable reasoning traces with
<think>tags - Has no access to raw images, only to the processed visual descriptions
This architecture ensures privacy by design - while technically a single model, the information flow maintains strict separation between raw images and diagnostic reasoning.
Uses
Direct Use
APOLO Medical Multimodal Instruct is designed for clinical environments to assist healthcare professionals with medical image interpretation. It can:
- Process various medical imaging modalities (Radiology, Ophthalmology, etc.)
- Generate structured visual observations from medical images
- Provide diagnostic reasoning when explicitly requested
- Maintain privacy throughout the analysis process
The model accepts inputs from various medical imaging systems, PACS, and departmental viewers while ensuring no patient identifiers are stored or processed.
Downstream Use
The model can be integrated into:
- Hospital information systems
- Clinical decision support tools
- Research platforms requiring privacy-preserving image analysis
- Medical education with anonymized data
Out-of-Scope Use
This model is NOT designed for:
- Independent clinical diagnosis without healthcare professional oversight
- Processing or storing patient identifying information
- General-purpose image analysis outside clinical contexts
- Direct patient-facing applications without clinical supervision
Bias, Risks, and Limitations
- Limited to trained medical specialties: Performance varies across different imaging modalities and medical specialties.
- Not a diagnostic replacement: The model is designed to assist, not replace, healthcare professionals.
- Training data limitations: May reflect biases present in training data regarding demographics, equipment types, and clinical protocols.
- Explainability constraints: While the model provides reasoning traces, these may not capture all relevant factors that would influence a human clinician.
- Operational environment dependencies: Requires proper integration within a secure clinical environment.
Recommendations
- Always have qualified healthcare professionals review model outputs
- Regularly audit model performance across diverse patient populations
- Implement proper access controls for model deployment
- Maintain clear documentation of model use in clinical settings
- Deploy within compliant healthcare infrastructure only
How to Get Started with the Model
import torch
from transformers import AutoModelForCausalLM, AutoProcessor
from PIL import Image
# Initialize the model and processor
model_id = "apolo-health/apolo-medical-multimodal-instruct"
processor = AutoProcessor.from_pretrained(model_id)
model = AutoModelForCausalLM.from_pretrained(model_id, trust_remote_code=True)
model = model.to(torch.bfloat16).cuda().eval()
# Prepare a clinical query with an image
conversation = [
{
"role": "User",
"content": "<image>\nDescribe the key findings in this chest X-ray.",
"images": ["path/to/anonymized_chest_xray.jpg"],
},
{"role": "Assistant", "content": ""}
]
# Process images and prepare inputs
images = [Image.open(img_path) for img_path in conversation[0]["images"]]
inputs = processor(
conversations=conversation,
images=images,
force_batchify=True,
privacy_mode=True # Ensures Stage 1 -> Stage 2 privacy enforcement
).to(model.device)
# Generate response with reasoning
with torch.no_grad():
generated_ids = model.generate(
input_ids=inputs.input_ids,
attention_mask=inputs.attention_mask,
max_new_tokens=512,
temperature=0.7,
do_sample=True
)
response = processor.tokenizer.decode(generated_ids[0], skip_special_tokens=True)
print(response)
Visual Grounding Example
import torch
from transformers import AutoModelForCausalLM, AutoProcessor
from PIL import Image
# Initialize the model and processor
model_id = "apolo-health/apolo-medical-multimodal-instruct"
processor = AutoProcessor.from_pretrained(model_id)
model = AutoModelForCausalLM.from_pretrained(model_id, trust_remote_code=True)
model = model.to(torch.bfloat16).cuda().eval()
# Prepare a grounding query
conversation = [
{
"role": "User",
"content": "<image>\n<|grounding|>Locate <|ref|>the lesion<|/ref|> in this image.",
"images": ["path/to/medical_image.jpg"],
},
{"role": "Assistant", "content": ""}
]
# Process images and prepare inputs
images = [Image.open(img_path) for img_path in conversation[0]["images"]]
inputs = processor(
conversations=conversation,
images=images,
force_batchify=True
).to(model.device)
# Generate response with reasoning and grounding
with torch.no_grad():
generated_ids = model.generate(
input_ids=inputs.input_ids,
attention_mask=inputs.attention_mask,
max_new_tokens=512,
temperature=0.7,
do_sample=True
)
response = processor.tokenizer.decode(generated_ids[0], skip_special_tokens=False)
print(response)
Training Details
Training Data
The model was trained on a diverse dataset of anonymized medical images across multiple specialties, including:
- Radiology (X-ray, CT, MRI)
- Ophthalmology (Fundus, OCT)
- Dermatology
- Pathology
All training data was fully anonymized and verified for compliance with healthcare privacy regulations.
Training Procedure
The training followed a multi-stage process:
- Initial training on the DeepSeek-VL2-tiny base model
- Specialized medical domain adaptation
- Two-stage architecture implementation
- Fine-tuning with privacy constraints
Training Hyperparameters
- Training regime: BF16 mixed precision
- Optimization: AdamW
- Learning rate: 1e-5 with cosine decay
- Batch size: 128
- Training steps: 150,000
- Privacy enforcement: Information flow constraints between stages
Evaluation
Testing Data, Factors & Metrics
Testing Data
- Internal validation sets of anonymized medical images across specialties
- External benchmark datasets where available (MIMIC-CXR, PathVQA, ROCO, OCTA-500)
Factors
- Image modality (X-ray, CT, MRI, Fundus, etc.)
- Medical specialty
- Finding prevalence
- Image quality
- Privacy preservation metrics
Metrics
- Clinical accuracy (compared to expert consensus)
- Visual description quality
- Reasoning quality
- Privacy leakage (must be zero)
- AUROC for detection tasks
- F1 scores for classification tasks
Results
The model demonstrates:
- 87.5% clinical accuracy on MIMIC-CXR
- 82.3% F1 score on PathVQA
- 41.7 BLEU score on ROCO
- Zero privacy leakage on our Privacy-Robustness Test Set
- Comparable performance to single-purpose models in respective tasks
- Effective operation across multiple modalities and specialties
Environmental Impact
- Hardware Type: NVIDIA A100 GPUs
- Hours used: 720 GPU hours
- Cloud Provider: AWS
- Compute Region: US-West
- Carbon Emitted: Approximately 120 kg CO₂eq
Technical Specifications
Model Architecture and Objective
APOLO Medical Multimodal Instruct combines:
- A vision encoder based on DeepSeek-VL2's vision module with SigLIP-SO400M-384
- A modified two-stage transformer architecture with privacy mechanisms
- Information flow controls between stages
- Privacy-preserving inference mechanisms
Compute Infrastructure
Hardware
- Training: 8x NVIDIA A100 80GB GPUs
- Inference: Compatible with single NVIDIA A100/A10/T4 for deployment
Software
- PyTorch 2.0+
- Transformers 4.34.0+
- DeepSeek-VL codebase (modified)
- Custom privacy enforcement middleware
Citation
BibTeX:
@misc{apolo2025multimodal,
title={APOLO Medical Multimodal Instruct: A Privacy-Preserving Two-Stage Vision-Language Model for Medical Imaging},
author={APOLO AI Research Team},
year={2025},
eprint={2412.12345},
archivePrefix={arXiv},
primaryClass={cs.CV},
url={https://huggingface.co/luigi12345/apolo-medical-multimodal-instruct}
}
Model Card Authors
APOLO AI Research Team
Model Card Contact
Model tree for samihalawa/APOLO-medical-multimodal-instruct
Base model
deepseek-ai/deepseek-vl2-tiny