YAML Metadata
Error:
"widget" must be an array
Model description
This is a LightGBM model trained on horse health outcome data from Kaggle.
Intended uses & limitations
This model is not ready to be used in production.
Training Procedure
[More Information Needed]
Hyperparameters
Click to expand
| Hyperparameter | Value |
|---|---|
| memory | |
| steps | [('preprocessor', ColumnTransformer(remainder='passthrough', transformers=[('num', Pipeline(steps=[('imputer', SimpleImputer(strategy='median')), ('scaler', StandardScaler())]), ['rectal_temp', 'pulse', 'respiratory_rate', 'nasogastric_reflux_ph', 'packed_cell_volume', 'total_protein', 'abdomo_protein', 'lesion_1', 'lesion_2', 'lesion_3']), ('cat', Pipeline(steps=[('imputer', SimpleI...='missing', strategy='constant')), ('onehot', OneHotEncoder(handle_unknown='ignore'))]), ['surgery', 'age', 'temp_of_extremities', 'peripheral_pulse', 'mucous_membrane', 'capillary_refill_time', 'pain', 'peristalsis', 'abdominal_distention', 'nasogastric_tube', 'nasogastric_reflux', 'rectal_exam_feces', 'abdomen', 'abdomo_appearance', 'surgical_lesion', 'cp_data'])])), ('classifier', LGBMClassifier(max_depth=3))] |
| verbose | False |
| preprocessor | ColumnTransformer(remainder='passthrough', transformers=[('num', Pipeline(steps=[('imputer', SimpleImputer(strategy='median')), ('scaler', StandardScaler())]), ['rectal_temp', 'pulse', 'respiratory_rate', 'nasogastric_reflux_ph', 'packed_cell_volume', 'total_protein', 'abdomo_protein', 'lesion_1', 'lesion_2', 'lesion_3']), ('cat', Pipeline(steps=[('imputer', SimpleI...='missing', strategy='constant')), ('onehot', OneHotEncoder(handle_unknown='ignore'))]), ['surgery', 'age', 'temp_of_extremities', 'peripheral_pulse', 'mucous_membrane', 'capillary_refill_time', 'pain', 'peristalsis', 'abdominal_distention', 'nasogastric_tube', 'nasogastric_reflux', 'rectal_exam_feces', 'abdomen', 'abdomo_appearance', 'surgical_lesion', 'cp_data'])]) |
| classifier | LGBMClassifier(max_depth=3) |
| preprocessor__n_jobs | |
| preprocessor__remainder | passthrough |
| preprocessor__sparse_threshold | 0.3 |
| preprocessor__transformer_weights | |
| preprocessor__transformers | [('num', Pipeline(steps=[('imputer', SimpleImputer(strategy='median')), ('scaler', StandardScaler())]), ['rectal_temp', 'pulse', 'respiratory_rate', 'nasogastric_reflux_ph', 'packed_cell_volume', 'total_protein', 'abdomo_protein', 'lesion_1', 'lesion_2', 'lesion_3']), ('cat', Pipeline(steps=[('imputer', SimpleImputer(fill_value='missing', strategy='constant')), ('onehot', OneHotEncoder(handle_unknown='ignore'))]), ['surgery', 'age', 'temp_of_extremities', 'peripheral_pulse', 'mucous_membrane', 'capillary_refill_time', 'pain', 'peristalsis', 'abdominal_distention', 'nasogastric_tube', 'nasogastric_reflux', 'rectal_exam_feces', 'abdomen', 'abdomo_appearance', 'surgical_lesion', 'cp_data'])] |
| preprocessor__verbose | False |
| preprocessor__verbose_feature_names_out | True |
| preprocessor__num | Pipeline(steps=[('imputer', SimpleImputer(strategy='median')), ('scaler', StandardScaler())]) |
| preprocessor__cat | Pipeline(steps=[('imputer', SimpleImputer(fill_value='missing', strategy='constant')), ('onehot', OneHotEncoder(handle_unknown='ignore'))]) |
| preprocessor__num__memory | |
| preprocessor__num__steps | [('imputer', SimpleImputer(strategy='median')), ('scaler', StandardScaler())] |
| preprocessor__num__verbose | False |
| preprocessor__num__imputer | SimpleImputer(strategy='median') |
| preprocessor__num__scaler | StandardScaler() |
| preprocessor__num__imputer__add_indicator | False |
| preprocessor__num__imputer__copy | True |
| preprocessor__num__imputer__fill_value | |
| preprocessor__num__imputer__keep_empty_features | False |
| preprocessor__num__imputer__missing_values | nan |
| preprocessor__num__imputer__strategy | median |
| preprocessor__num__scaler__copy | True |
| preprocessor__num__scaler__with_mean | True |
| preprocessor__num__scaler__with_std | True |
| preprocessor__cat__memory | |
| preprocessor__cat__steps | [('imputer', SimpleImputer(fill_value='missing', strategy='constant')), ('onehot', OneHotEncoder(handle_unknown='ignore'))] |
| preprocessor__cat__verbose | False |
| preprocessor__cat__imputer | SimpleImputer(fill_value='missing', strategy='constant') |
| preprocessor__cat__onehot | OneHotEncoder(handle_unknown='ignore') |
| preprocessor__cat__imputer__add_indicator | False |
| preprocessor__cat__imputer__copy | True |
| preprocessor__cat__imputer__fill_value | missing |
| preprocessor__cat__imputer__keep_empty_features | False |
| preprocessor__cat__imputer__missing_values | nan |
| preprocessor__cat__imputer__strategy | constant |
| preprocessor__cat__onehot__categories | auto |
| preprocessor__cat__onehot__drop | |
| preprocessor__cat__onehot__dtype | <class 'numpy.float64'> |
| preprocessor__cat__onehot__feature_name_combiner | concat |
| preprocessor__cat__onehot__handle_unknown | ignore |
| preprocessor__cat__onehot__max_categories | |
| preprocessor__cat__onehot__min_frequency | |
| preprocessor__cat__onehot__sparse | deprecated |
| preprocessor__cat__onehot__sparse_output | True |
| classifier__boosting_type | gbdt |
| classifier__class_weight | |
| classifier__colsample_bytree | 1.0 |
| classifier__importance_type | split |
| classifier__learning_rate | 0.1 |
| classifier__max_depth | 3 |
| classifier__min_child_samples | 20 |
| classifier__min_child_weight | 0.001 |
| classifier__min_split_gain | 0.0 |
| classifier__n_estimators | 100 |
| classifier__n_jobs | |
| classifier__num_leaves | 31 |
| classifier__objective | |
| classifier__random_state | |
| classifier__reg_alpha | 0.0 |
| classifier__reg_lambda | 0.0 |
| classifier__subsample | 1.0 |
| classifier__subsample_for_bin | 200000 |
| classifier__subsample_freq | 0 |
Model Plot
Pipeline(steps=[('preprocessor',ColumnTransformer(remainder='passthrough',transformers=[('num',Pipeline(steps=[('imputer',SimpleImputer(strategy='median')),('scaler',StandardScaler())]),['rectal_temp', 'pulse','respiratory_rate','nasogastric_reflux_ph','packed_cell_volume','total_protein','abdomo_protein', 'lesion_1','lesion_2', 'lesion_3']),('cat',Pi...OneHotEncoder(handle_unknown='ignore'))]),['surgery', 'age','temp_of_extremities','peripheral_pulse','mucous_membrane','capillary_refill_time','pain', 'peristalsis','abdominal_distention','nasogastric_tube','nasogastric_reflux','rectal_exam_feces','abdomen','abdomo_appearance','surgical_lesion','cp_data'])])),('classifier', LGBMClassifier(max_depth=3))])In a Jupyter environment, please rerun this cell to show the HTML representation or trust the notebook. On GitHub, the HTML representation is unable to render, please try loading this page with nbviewer.org.
Pipeline(steps=[('preprocessor',ColumnTransformer(remainder='passthrough',transformers=[('num',Pipeline(steps=[('imputer',SimpleImputer(strategy='median')),('scaler',StandardScaler())]),['rectal_temp', 'pulse','respiratory_rate','nasogastric_reflux_ph','packed_cell_volume','total_protein','abdomo_protein', 'lesion_1','lesion_2', 'lesion_3']),('cat',Pi...OneHotEncoder(handle_unknown='ignore'))]),['surgery', 'age','temp_of_extremities','peripheral_pulse','mucous_membrane','capillary_refill_time','pain', 'peristalsis','abdominal_distention','nasogastric_tube','nasogastric_reflux','rectal_exam_feces','abdomen','abdomo_appearance','surgical_lesion','cp_data'])])),('classifier', LGBMClassifier(max_depth=3))])ColumnTransformer(remainder='passthrough',transformers=[('num',Pipeline(steps=[('imputer',SimpleImputer(strategy='median')),('scaler', StandardScaler())]),['rectal_temp', 'pulse', 'respiratory_rate','nasogastric_reflux_ph', 'packed_cell_volume','total_protein', 'abdomo_protein', 'lesion_1','lesion_2', 'lesion_3']),('cat',Pipeline(steps=[('imputer',SimpleI...='missing',strategy='constant')),('onehot',OneHotEncoder(handle_unknown='ignore'))]),['surgery', 'age', 'temp_of_extremities','peripheral_pulse', 'mucous_membrane','capillary_refill_time', 'pain','peristalsis', 'abdominal_distention','nasogastric_tube', 'nasogastric_reflux','rectal_exam_feces', 'abdomen','abdomo_appearance', 'surgical_lesion','cp_data'])])['rectal_temp', 'pulse', 'respiratory_rate', 'nasogastric_reflux_ph', 'packed_cell_volume', 'total_protein', 'abdomo_protein', 'lesion_1', 'lesion_2', 'lesion_3']
SimpleImputer(strategy='median')
StandardScaler()
['surgery', 'age', 'temp_of_extremities', 'peripheral_pulse', 'mucous_membrane', 'capillary_refill_time', 'pain', 'peristalsis', 'abdominal_distention', 'nasogastric_tube', 'nasogastric_reflux', 'rectal_exam_feces', 'abdomen', 'abdomo_appearance', 'surgical_lesion', 'cp_data']
SimpleImputer(fill_value='missing', strategy='constant')
OneHotEncoder(handle_unknown='ignore')
[]
passthrough
LGBMClassifier(max_depth=3)
Evaluation Results
| Metric | Value |
|---|---|
| accuracy | 0.740891 |
| f1 score | 0.740891 |
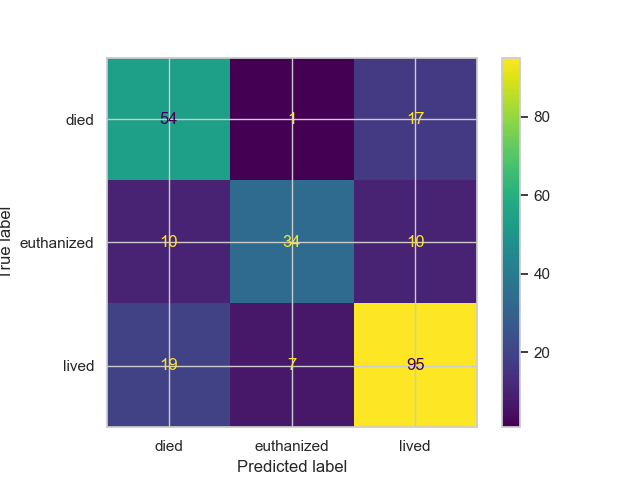
Confusion Matrix
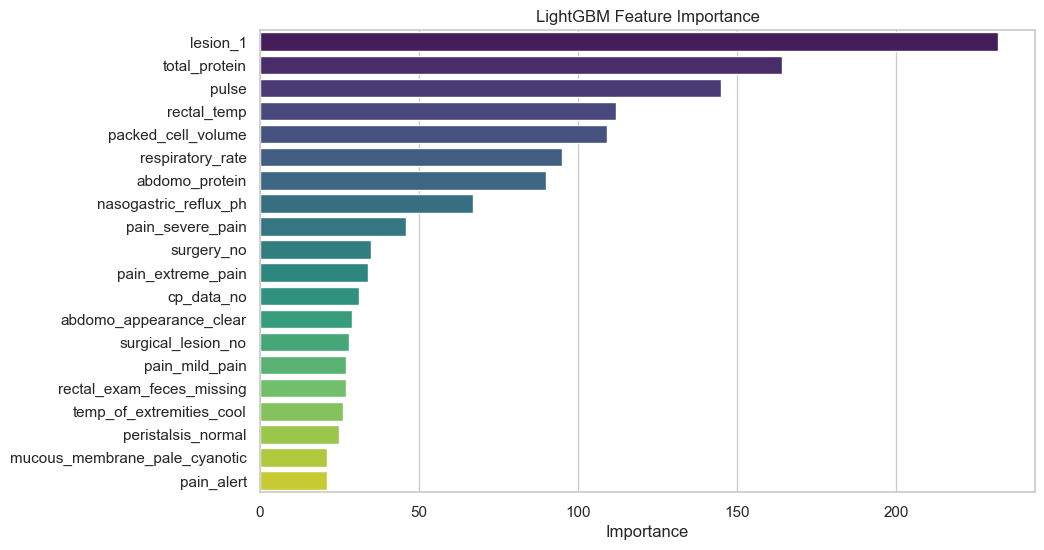
Permutation Importance
How to Get Started with the Model
[More Information Needed]
Model Card Authors
kmposkid
Model Card Contact
You can contact the model card authors through following channels: [More Information Needed]
Citation
Below you can find information related to citation.
BibTeX:
[More Information Needed]
- Downloads last month
- 0
Inference Providers
NEW
This model isn't deployed by any Inference Provider.
🙋
Ask for provider support