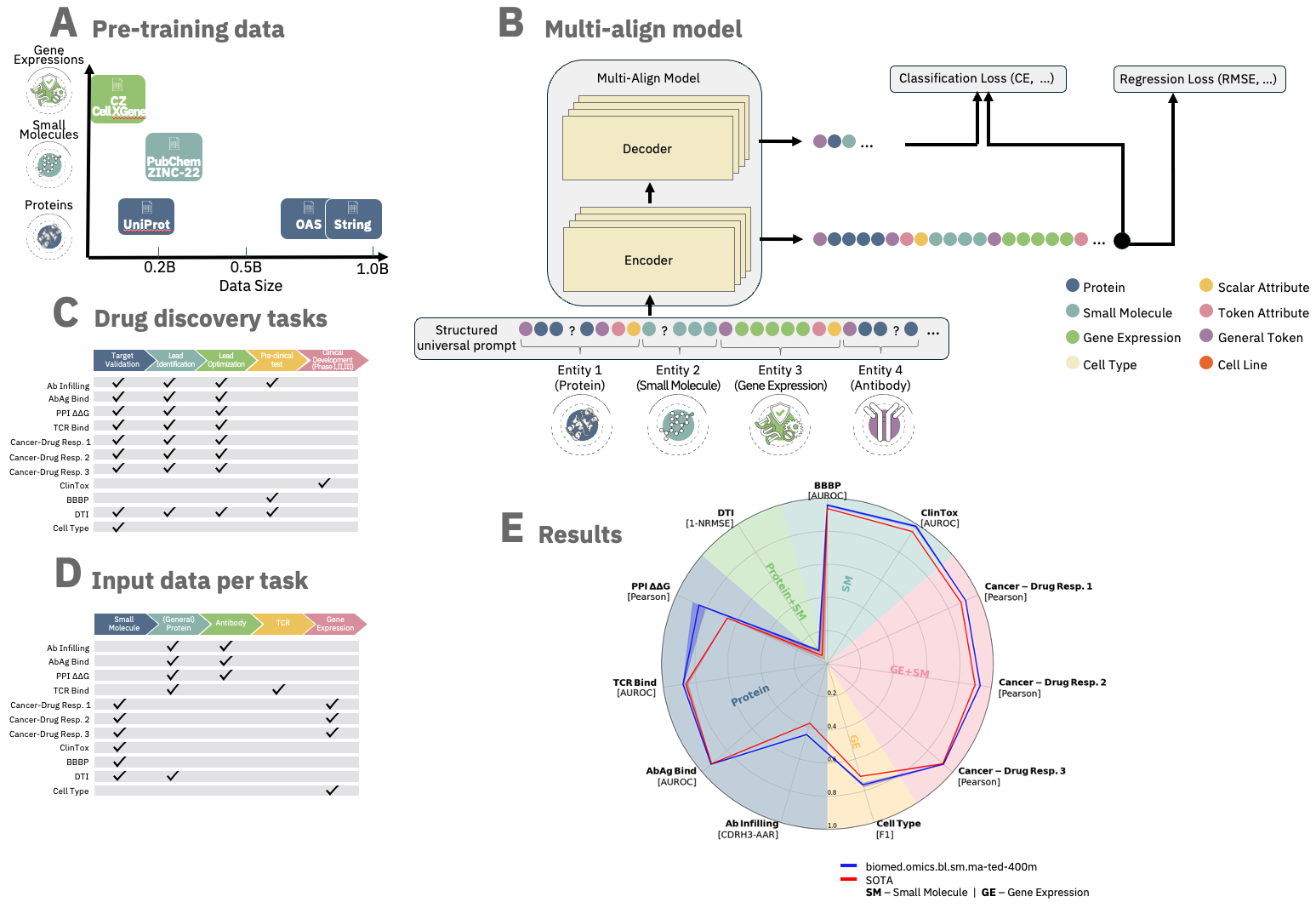
The ibm/biomed.omics.bl.sm.ma-ted-458m model is a biomedical foundation model trained on over 2 billion biological samples across multiple modalities, including proteins, small molecules, and single-cell gene data.
Designed for robust performance, it achieves state-of-the-art results over a variety of tasks across the entire drug discovery pipeline and the diverse biomedical domains.
Based on the Molecular Aligned Multi-Modal Architecture and Language (MAMMAL), a flexible, multi-domain architecture with an adaptable task prompt syntax.
The syntax allows for dynamic combinations of tokens and scalars, enabling classification, regression, and generation tasks either within a single domain or with cross-domain entities.
Model Summary
- Developers: IBM Research
- GitHub Repository: https://github.com/BiomedSciAI/biomed-multi-alignment
- Paper: https://arxiv.org/abs/2410.22367
- Release Date: Oct 28th, 2024
- License: Apache 2.0.
Usage
Using ibm/biomed.omics.bl.sm.ma-ted-458m requires installing https://github.com/BiomedSciAI/biomed-multi-alignment
pip install git+https://github.com/BiomedSciAI/biomed-multi-alignment.git
A simple example for a task already supported by ibm/biomed.omics.bl.sm.ma-ted-458m:
import torch
from fuse.data.tokenizers.modular_tokenizer.op import ModularTokenizerOp
from mammal.model import Mammal
from mammal.keys import *
# Load Model
model = Mammal.from_pretrained("ibm/biomed.omics.bl.sm.ma-ted-458m")
model.eval()
# Load Tokenizer
tokenizer_op = ModularTokenizerOp.from_pretrained("ibm/biomed.omics.bl.sm.ma-ted-458m")
# Prepare Input Prompt
protein_calmodulin = "MADQLTEEQIAEFKEAFSLFDKDGDGTITTKELGTVMRSLGQNPTEAELQDMISELDQDGFIDKEDLHDGDGKISFEEFLNLVNKEMTADVDGDGQVNYEEFVTMMTSK"
protein_calcineurin = "MSSKLLLAGLDIERVLAEKNFYKEWDTWIIEAMNVGDEEVDRIKEFKEDEIFEEAKTLGTAEMQEYKKQKLEEAIEGAFDIFDKDGNGYISAAELRHVMTNLGEKLTDEEVDEMIRQMWDQNGDWDRIKELKFGEIKKLSAKDTRGTIFIKVFENLGTGVDSEYEDVSKYMLKHQ"
# Create and load sample
sample_dict = dict()
# Formatting prompt to match pre-training syntax
sample_dict[ENCODER_INPUTS_STR] = f"<@TOKENIZER-TYPE=AA><BINDING_AFFINITY_CLASS><SENTINEL_ID_0><MOLECULAR_ENTITY><MOLECULAR_ENTITY_GENERAL_PROTEIN><SEQUENCE_NATURAL_START>{protein_calmodulin}<SEQUENCE_NATURAL_END><MOLECULAR_ENTITY><MOLECULAR_ENTITY_GENERAL_PROTEIN><SEQUENCE_NATURAL_START>{protein_calcineurin}<SEQUENCE_NATURAL_END><EOS>"
# Tokenize
tokenizer_op(
sample_dict=sample_dict,
key_in=ENCODER_INPUTS_STR,
key_out_tokens_ids=ENCODER_INPUTS_TOKENS,
key_out_attention_mask=ENCODER_INPUTS_ATTENTION_MASK,
)
sample_dict[ENCODER_INPUTS_TOKENS] = torch.tensor(sample_dict[ENCODER_INPUTS_TOKENS])
sample_dict[ENCODER_INPUTS_ATTENTION_MASK] = torch.tensor(sample_dict[ENCODER_INPUTS_ATTENTION_MASK])
# Generate Prediction
batch_dict = model.generate(
[sample_dict],
output_scores=True,
return_dict_in_generate=True,
max_new_tokens=5,
)
# Get output
generated_output = tokenizer_op._tokenizer.decode(batch_dict[CLS_PRED][0])
print(f"{generated_output=}")
For more advanced usage, see our detailed example at:
Citation
If you found our work useful, please consider to give a star to the repo and cite our paper:
@misc{shoshan2024mammalmolecularaligned,
title={MAMMAL -- Molecular Aligned Multi-Modal Architecture and Language},
author={Yoel Shoshan and Moshiko Raboh and Michal Ozery-Flato and Vadim Ratner and Alex Golts and Jeffrey K. Weber and Ella Barkan and Simona Rabinovici-Cohen and Sagi Polaczek and Ido Amos and Ben Shapira and Liam Hazan and Matan Ninio and Sivan Ravid and Michael M. Danziger and Joseph A. Morrone and Parthasarathy Suryanarayanan and Michal Rosen-Zvi and Efrat Hexter},
year={2024},
eprint={2410.22367},
archivePrefix={arXiv},
primaryClass={q-bio.QM},
url={https://arxiv.org/abs/2410.22367},
}
- Downloads last month
- 363