__key__
stringlengths 32
39
| stainer
stringclasses 1
value | scanner
stringclasses 4
values | slide_id
stringclasses 4
values | tile_id
stringlengths 12
15
| png
imagewidth (px) 224
224
|
|---|---|---|---|---|---|
./GIVH_AT2_to_GMH_S60/tile_16_100_127 | GIVH | AT2 | GIVH_AT2_to_GMH_S60 | tile_16_100_127 | |
./GIVH_AT2_to_GMH_S60/tile_16_100_128 | GIVH | AT2 | GIVH_AT2_to_GMH_S60 | tile_16_100_128 | |
./GIVH_AT2_to_GMH_S60/tile_16_100_129 | GIVH | AT2 | GIVH_AT2_to_GMH_S60 | tile_16_100_129 | |
./GIVH_AT2_to_GMH_S60/tile_16_100_130 | GIVH | AT2 | GIVH_AT2_to_GMH_S60 | tile_16_100_130 | |
./GIVH_AT2_to_GMH_S60/tile_16_100_131 | GIVH | AT2 | GIVH_AT2_to_GMH_S60 | tile_16_100_131 | |
./GIVH_AT2_to_GMH_S60/tile_16_100_132 | GIVH | AT2 | GIVH_AT2_to_GMH_S60 | tile_16_100_132 | |
./GIVH_AT2_to_GMH_S60/tile_16_100_133 | GIVH | AT2 | GIVH_AT2_to_GMH_S60 | tile_16_100_133 | |
./GIVH_AT2_to_GMH_S60/tile_16_100_134 | GIVH | AT2 | GIVH_AT2_to_GMH_S60 | tile_16_100_134 | |
./GIVH_AT2_to_GMH_S60/tile_16_100_135 | GIVH | AT2 | GIVH_AT2_to_GMH_S60 | tile_16_100_135 | |
./GIVH_AT2_to_GMH_S60/tile_16_100_136 | GIVH | AT2 | GIVH_AT2_to_GMH_S60 | tile_16_100_136 | |
./GIVH_AT2_to_GMH_S60/tile_16_100_137 | GIVH | AT2 | GIVH_AT2_to_GMH_S60 | tile_16_100_137 | |
./GIVH_AT2_to_GMH_S60/tile_16_101_126 | GIVH | AT2 | GIVH_AT2_to_GMH_S60 | tile_16_101_126 | |
./GIVH_AT2_to_GMH_S60/tile_16_101_127 | GIVH | AT2 | GIVH_AT2_to_GMH_S60 | tile_16_101_127 | |
./GIVH_AT2_to_GMH_S60/tile_16_101_128 | GIVH | AT2 | GIVH_AT2_to_GMH_S60 | tile_16_101_128 | |
./GIVH_AT2_to_GMH_S60/tile_16_101_129 | GIVH | AT2 | GIVH_AT2_to_GMH_S60 | tile_16_101_129 | |
./GIVH_AT2_to_GMH_S60/tile_16_101_130 | GIVH | AT2 | GIVH_AT2_to_GMH_S60 | tile_16_101_130 | |
./GIVH_AT2_to_GMH_S60/tile_16_101_131 | GIVH | AT2 | GIVH_AT2_to_GMH_S60 | tile_16_101_131 | |
./GIVH_AT2_to_GMH_S60/tile_16_101_132 | GIVH | AT2 | GIVH_AT2_to_GMH_S60 | tile_16_101_132 | |
./GIVH_AT2_to_GMH_S60/tile_16_101_133 | GIVH | AT2 | GIVH_AT2_to_GMH_S60 | tile_16_101_133 | |
./GIVH_AT2_to_GMH_S60/tile_16_101_134 | GIVH | AT2 | GIVH_AT2_to_GMH_S60 | tile_16_101_134 | |
./GIVH_AT2_to_GMH_S60/tile_16_101_135 | GIVH | AT2 | GIVH_AT2_to_GMH_S60 | tile_16_101_135 | |
./GIVH_AT2_to_GMH_S60/tile_16_101_136 | GIVH | AT2 | GIVH_AT2_to_GMH_S60 | tile_16_101_136 | |
./GIVH_AT2_to_GMH_S60/tile_16_101_137 | GIVH | AT2 | GIVH_AT2_to_GMH_S60 | tile_16_101_137 | |
./GIVH_AT2_to_GMH_S60/tile_16_101_138 | GIVH | AT2 | GIVH_AT2_to_GMH_S60 | tile_16_101_138 | |
./GIVH_AT2_to_GMH_S60/tile_16_101_25 | GIVH | AT2 | GIVH_AT2_to_GMH_S60 | tile_16_101_25 | |
./GIVH_AT2_to_GMH_S60/tile_16_101_26 | GIVH | AT2 | GIVH_AT2_to_GMH_S60 | tile_16_101_26 | |
./GIVH_AT2_to_GMH_S60/tile_16_101_27 | GIVH | AT2 | GIVH_AT2_to_GMH_S60 | tile_16_101_27 | |
./GIVH_AT2_to_GMH_S60/tile_16_102_124 | GIVH | AT2 | GIVH_AT2_to_GMH_S60 | tile_16_102_124 | |
./GIVH_AT2_to_GMH_S60/tile_16_102_126 | GIVH | AT2 | GIVH_AT2_to_GMH_S60 | tile_16_102_126 | |
./GIVH_AT2_to_GMH_S60/tile_16_102_127 | GIVH | AT2 | GIVH_AT2_to_GMH_S60 | tile_16_102_127 | |
./GIVH_AT2_to_GMH_S60/tile_16_102_128 | GIVH | AT2 | GIVH_AT2_to_GMH_S60 | tile_16_102_128 | |
./GIVH_AT2_to_GMH_S60/tile_16_102_129 | GIVH | AT2 | GIVH_AT2_to_GMH_S60 | tile_16_102_129 | |
./GIVH_AT2_to_GMH_S60/tile_16_102_130 | GIVH | AT2 | GIVH_AT2_to_GMH_S60 | tile_16_102_130 | |
./GIVH_AT2_to_GMH_S60/tile_16_102_131 | GIVH | AT2 | GIVH_AT2_to_GMH_S60 | tile_16_102_131 | |
./GIVH_AT2_to_GMH_S60/tile_16_102_132 | GIVH | AT2 | GIVH_AT2_to_GMH_S60 | tile_16_102_132 | |
./GIVH_AT2_to_GMH_S60/tile_16_102_133 | GIVH | AT2 | GIVH_AT2_to_GMH_S60 | tile_16_102_133 | |
./GIVH_AT2_to_GMH_S60/tile_16_102_134 | GIVH | AT2 | GIVH_AT2_to_GMH_S60 | tile_16_102_134 | |
./GIVH_AT2_to_GMH_S60/tile_16_102_135 | GIVH | AT2 | GIVH_AT2_to_GMH_S60 | tile_16_102_135 | |
./GIVH_AT2_to_GMH_S60/tile_16_102_136 | GIVH | AT2 | GIVH_AT2_to_GMH_S60 | tile_16_102_136 | |
./GIVH_AT2_to_GMH_S60/tile_16_102_137 | GIVH | AT2 | GIVH_AT2_to_GMH_S60 | tile_16_102_137 | |
./GIVH_AT2_to_GMH_S60/tile_16_102_138 | GIVH | AT2 | GIVH_AT2_to_GMH_S60 | tile_16_102_138 | |
./GIVH_AT2_to_GMH_S60/tile_16_102_139 | GIVH | AT2 | GIVH_AT2_to_GMH_S60 | tile_16_102_139 | |
./GIVH_AT2_to_GMH_S60/tile_16_102_140 | GIVH | AT2 | GIVH_AT2_to_GMH_S60 | tile_16_102_140 | |
./GIVH_AT2_to_GMH_S60/tile_16_103_124 | GIVH | AT2 | GIVH_AT2_to_GMH_S60 | tile_16_103_124 | |
./GIVH_AT2_to_GMH_S60/tile_16_103_125 | GIVH | AT2 | GIVH_AT2_to_GMH_S60 | tile_16_103_125 | |
./GIVH_AT2_to_GMH_S60/tile_16_103_126 | GIVH | AT2 | GIVH_AT2_to_GMH_S60 | tile_16_103_126 | |
./GIVH_AT2_to_GMH_S60/tile_16_103_127 | GIVH | AT2 | GIVH_AT2_to_GMH_S60 | tile_16_103_127 | |
./GIVH_AT2_to_GMH_S60/tile_16_103_128 | GIVH | AT2 | GIVH_AT2_to_GMH_S60 | tile_16_103_128 | |
./GIVH_AT2_to_GMH_S60/tile_16_103_129 | GIVH | AT2 | GIVH_AT2_to_GMH_S60 | tile_16_103_129 | |
./GIVH_AT2_to_GMH_S60/tile_16_103_130 | GIVH | AT2 | GIVH_AT2_to_GMH_S60 | tile_16_103_130 | |
./GIVH_AT2_to_GMH_S60/tile_16_103_131 | GIVH | AT2 | GIVH_AT2_to_GMH_S60 | tile_16_103_131 | |
./GIVH_AT2_to_GMH_S60/tile_16_103_132 | GIVH | AT2 | GIVH_AT2_to_GMH_S60 | tile_16_103_132 | |
./GIVH_AT2_to_GMH_S60/tile_16_103_133 | GIVH | AT2 | GIVH_AT2_to_GMH_S60 | tile_16_103_133 | |
./GIVH_AT2_to_GMH_S60/tile_16_103_134 | GIVH | AT2 | GIVH_AT2_to_GMH_S60 | tile_16_103_134 | |
./GIVH_AT2_to_GMH_S60/tile_16_103_135 | GIVH | AT2 | GIVH_AT2_to_GMH_S60 | tile_16_103_135 | |
./GIVH_AT2_to_GMH_S60/tile_16_103_136 | GIVH | AT2 | GIVH_AT2_to_GMH_S60 | tile_16_103_136 | |
./GIVH_AT2_to_GMH_S60/tile_16_103_137 | GIVH | AT2 | GIVH_AT2_to_GMH_S60 | tile_16_103_137 | |
./GIVH_AT2_to_GMH_S60/tile_16_103_138 | GIVH | AT2 | GIVH_AT2_to_GMH_S60 | tile_16_103_138 | |
./GIVH_AT2_to_GMH_S60/tile_16_103_139 | GIVH | AT2 | GIVH_AT2_to_GMH_S60 | tile_16_103_139 | |
./GIVH_AT2_to_GMH_S60/tile_16_103_140 | GIVH | AT2 | GIVH_AT2_to_GMH_S60 | tile_16_103_140 | |
./GIVH_AT2_to_GMH_S60/tile_16_103_141 | GIVH | AT2 | GIVH_AT2_to_GMH_S60 | tile_16_103_141 | |
./GIVH_AT2_to_GMH_S60/tile_16_104_123 | GIVH | AT2 | GIVH_AT2_to_GMH_S60 | tile_16_104_123 | |
./GIVH_AT2_to_GMH_S60/tile_16_104_124 | GIVH | AT2 | GIVH_AT2_to_GMH_S60 | tile_16_104_124 | |
./GIVH_AT2_to_GMH_S60/tile_16_104_126 | GIVH | AT2 | GIVH_AT2_to_GMH_S60 | tile_16_104_126 | |
./GIVH_AT2_to_GMH_S60/tile_16_104_127 | GIVH | AT2 | GIVH_AT2_to_GMH_S60 | tile_16_104_127 | |
./GIVH_AT2_to_GMH_S60/tile_16_104_128 | GIVH | AT2 | GIVH_AT2_to_GMH_S60 | tile_16_104_128 | |
./GIVH_AT2_to_GMH_S60/tile_16_104_129 | GIVH | AT2 | GIVH_AT2_to_GMH_S60 | tile_16_104_129 | |
./GIVH_AT2_to_GMH_S60/tile_16_104_130 | GIVH | AT2 | GIVH_AT2_to_GMH_S60 | tile_16_104_130 | |
./GIVH_AT2_to_GMH_S60/tile_16_104_131 | GIVH | AT2 | GIVH_AT2_to_GMH_S60 | tile_16_104_131 | |
./GIVH_AT2_to_GMH_S60/tile_16_104_132 | GIVH | AT2 | GIVH_AT2_to_GMH_S60 | tile_16_104_132 | |
./GIVH_AT2_to_GMH_S60/tile_16_104_133 | GIVH | AT2 | GIVH_AT2_to_GMH_S60 | tile_16_104_133 | |
./GIVH_AT2_to_GMH_S60/tile_16_104_134 | GIVH | AT2 | GIVH_AT2_to_GMH_S60 | tile_16_104_134 | |
./GIVH_AT2_to_GMH_S60/tile_16_104_135 | GIVH | AT2 | GIVH_AT2_to_GMH_S60 | tile_16_104_135 | |
./GIVH_AT2_to_GMH_S60/tile_16_104_136 | GIVH | AT2 | GIVH_AT2_to_GMH_S60 | tile_16_104_136 | |
./GIVH_AT2_to_GMH_S60/tile_16_104_137 | GIVH | AT2 | GIVH_AT2_to_GMH_S60 | tile_16_104_137 | |
./GIVH_AT2_to_GMH_S60/tile_16_104_138 | GIVH | AT2 | GIVH_AT2_to_GMH_S60 | tile_16_104_138 | |
./GIVH_AT2_to_GMH_S60/tile_16_104_139 | GIVH | AT2 | GIVH_AT2_to_GMH_S60 | tile_16_104_139 | |
./GIVH_AT2_to_GMH_S60/tile_16_104_170 | GIVH | AT2 | GIVH_AT2_to_GMH_S60 | tile_16_104_170 | |
./GIVH_AT2_to_GMH_S60/tile_16_104_79 | GIVH | AT2 | GIVH_AT2_to_GMH_S60 | tile_16_104_79 | |
./GIVH_AT2_to_GMH_S60/tile_16_104_80 | GIVH | AT2 | GIVH_AT2_to_GMH_S60 | tile_16_104_80 | |
./GIVH_AT2_to_GMH_S60/tile_16_104_81 | GIVH | AT2 | GIVH_AT2_to_GMH_S60 | tile_16_104_81 | |
./GIVH_AT2_to_GMH_S60/tile_16_104_82 | GIVH | AT2 | GIVH_AT2_to_GMH_S60 | tile_16_104_82 | |
./GIVH_AT2_to_GMH_S60/tile_16_105_123 | GIVH | AT2 | GIVH_AT2_to_GMH_S60 | tile_16_105_123 | |
./GIVH_AT2_to_GMH_S60/tile_16_105_126 | GIVH | AT2 | GIVH_AT2_to_GMH_S60 | tile_16_105_126 | |
./GIVH_AT2_to_GMH_S60/tile_16_105_127 | GIVH | AT2 | GIVH_AT2_to_GMH_S60 | tile_16_105_127 | |
./GIVH_AT2_to_GMH_S60/tile_16_105_128 | GIVH | AT2 | GIVH_AT2_to_GMH_S60 | tile_16_105_128 | |
./GIVH_AT2_to_GMH_S60/tile_16_105_129 | GIVH | AT2 | GIVH_AT2_to_GMH_S60 | tile_16_105_129 | |
./GIVH_AT2_to_GMH_S60/tile_16_105_130 | GIVH | AT2 | GIVH_AT2_to_GMH_S60 | tile_16_105_130 | |
./GIVH_AT2_to_GMH_S60/tile_16_105_131 | GIVH | AT2 | GIVH_AT2_to_GMH_S60 | tile_16_105_131 | |
./GIVH_AT2_to_GMH_S60/tile_16_105_132 | GIVH | AT2 | GIVH_AT2_to_GMH_S60 | tile_16_105_132 | |
./GIVH_AT2_to_GMH_S60/tile_16_105_133 | GIVH | AT2 | GIVH_AT2_to_GMH_S60 | tile_16_105_133 | |
./GIVH_AT2_to_GMH_S60/tile_16_105_134 | GIVH | AT2 | GIVH_AT2_to_GMH_S60 | tile_16_105_134 | |
./GIVH_AT2_to_GMH_S60/tile_16_105_135 | GIVH | AT2 | GIVH_AT2_to_GMH_S60 | tile_16_105_135 | |
./GIVH_AT2_to_GMH_S60/tile_16_105_136 | GIVH | AT2 | GIVH_AT2_to_GMH_S60 | tile_16_105_136 | |
./GIVH_AT2_to_GMH_S60/tile_16_105_137 | GIVH | AT2 | GIVH_AT2_to_GMH_S60 | tile_16_105_137 | |
./GIVH_AT2_to_GMH_S60/tile_16_105_138 | GIVH | AT2 | GIVH_AT2_to_GMH_S60 | tile_16_105_138 | |
./GIVH_AT2_to_GMH_S60/tile_16_105_139 | GIVH | AT2 | GIVH_AT2_to_GMH_S60 | tile_16_105_139 | |
./GIVH_AT2_to_GMH_S60/tile_16_105_169 | GIVH | AT2 | GIVH_AT2_to_GMH_S60 | tile_16_105_169 | |
./GIVH_AT2_to_GMH_S60/tile_16_105_170 | GIVH | AT2 | GIVH_AT2_to_GMH_S60 | tile_16_105_170 | |
./GIVH_AT2_to_GMH_S60/tile_16_105_171 | GIVH | AT2 | GIVH_AT2_to_GMH_S60 | tile_16_105_171 |
PLISM dataset
The Pathology Images of Scanners and Mobilephones (PLISM) dataset was created by (Ochi et al., 2024) for the evaluation of AI models’ robustness to inter-institutional domain shifts. All histopathological specimens used in creating the PLISM dataset were sourced from patients who were diagnosed and underwent surgery at the University of Tokyo Hospital between 1955 and 2018.
PLISM-wsi consists in a group of consecutive slides digitized under 7 different scanners and stained accross 13 H&E conditions.
Each of the 91 sample encompasses the same biological information, that is a collection of 46 TMAs (Tissue Micro Arrays) from various organs. Additional details can be found in https://p024eb.github.io/ and the original publication
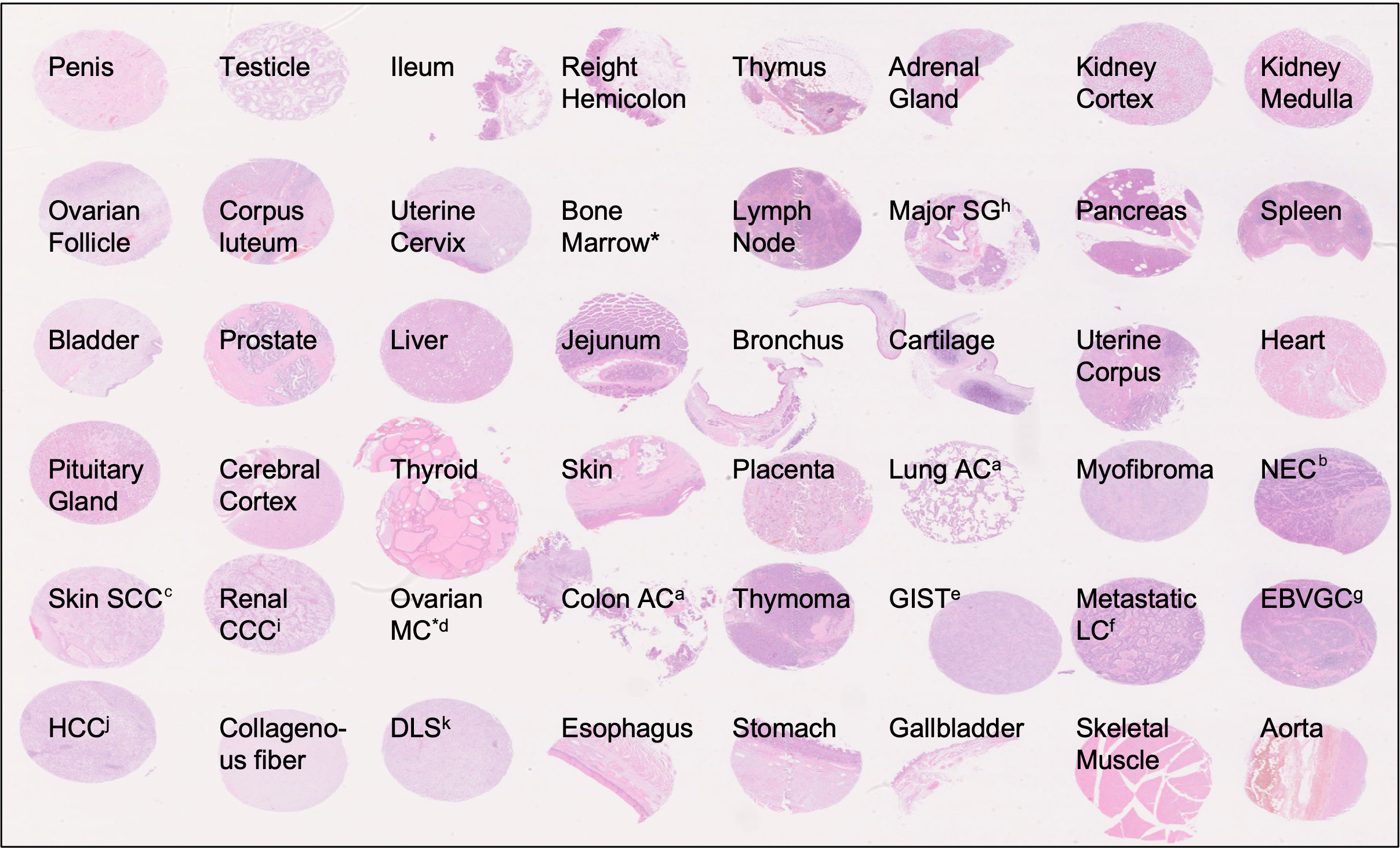
Figure 1: Tissue types included in TMA specimens of the PLISM-wsi dataset. Source: https://p024eb.github.io/ (Ochi et al., 2024)
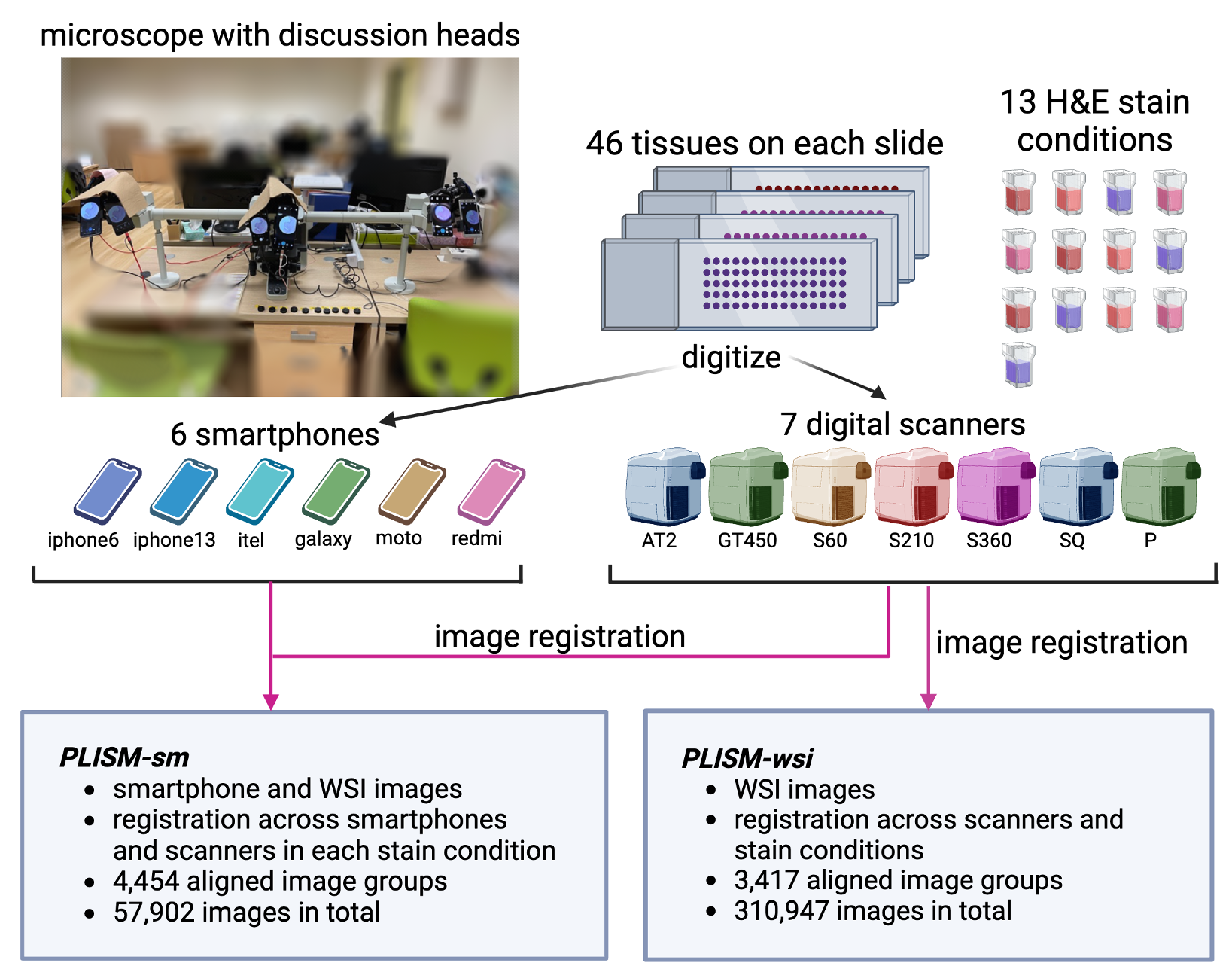
Figure 2: Digitization and staining worflow for PLISM dataset. Source: https://p024eb.github.io/ (Ochi et al., 2024)
PLISM dataset tiles
The original PLISM-wsi subset contains a total of 310,947 images.
Registration was performed across all scanners and staining conditions using OpenCV's AKAZE (Alcantarilla et al., 2013) key-point matching algorithm.
There were 3,417 aligned image groups, with a total of 310,947 (3,417 groups × 91 WSIs) image patches of shape 512x512 at a resolution ranging from 0.22 to 0.26 µm/pixel (40x magnification).
To follow the spirit of this unique and outstanding contribution, we generated an extended version of the original tiles dataset provided by (Ochi et al. 2024) so as to ease its adoption accross the digital pathology community and serve as a reference dataset for benchmarking the robustess of foundation models to staining and scanner variations. In particular, our work differs from the original dataset in the following aspects:
• The original, non-registered WSIs were registered using Elastix (Klein et al., 2010; Shamonin et al., 2014). The reference slide was stained with GMH condition and digitized using Hamamatsu Nanozoomer S60 scanner.
• Tiles of 224x224 pixels were extracted at mpp 0.5 µm/pixel (20x magnification) using an in-house bidirectionnal U-Net (Ronneberger et al., 2015).
• All tiles from the original WSI were extracted, resulting in 16,278 tiles for each of the 91 WSIs.
In total, our dataset encompasses 1,481,298 histology tiles for a total size of 150 Gb.
For each tile, we provide the original slide id (slide_id), tile id (tile_id), stainer and scanner.
How to extract features
🎉 Check plismbench to perform the feature extraction of PLISM dataset and get run our robustness benchmark 🎉
2h30 and roughly 10 Gb storage are necessary to extract all features with a ViT-B model, 16 CPUs and 1 Nvidia T4 (16Go).
License
This dataset is licensed under CC BY 4.0 licence.
Acknowledgments
We thank PLISM dataset's authors for their unique contribution.
Third-party licenses
- PLISM dataset (Ochi et al., 2024) is distributed under CC BY 4.0 license.
- Elastix (Klein et al., 2010; Shamonin et al., 2014) is distributed under Apache 2.0 license.
How to cite
If you are using this dataset, please cite the original article (Ochi et al., 2024) and our work as follows:
APA style
Filiot, A., Dop, N., Tchita, O., Riou, A., Peeters, T., Valter, D., Scalbert, M., Saillard, C., Robin, G., & Olivier, A. (2025). Distilling foundation models for robust and efficient models in digital pathology. arXiv. https://arxiv.org/abs/2501.16239
BibTex entry
@misc{filiot2025distillingfoundationmodelsrobust,
title={Distilling foundation models for robust and efficient models in digital pathology},
author={Alexandre Filiot and Nicolas Dop and Oussama Tchita and Auriane Riou and Thomas Peeters and Daria Valter and Marin Scalbert and Charlie Saillard and Geneviève Robin and Antoine Olivier},
year={2025},
eprint={2501.16239},
archivePrefix={arXiv},
primaryClass={cs.CV},
url={https://arxiv.org/abs/2501.16239},
}
References
(Ochi et al., 2024) Ochi, M., Komura, D., Onoyama, T. et al. Registered multi-device/staining histology image dataset for domain-agnostic machine learning models. Sci Data 11, 330 (2024).
(Alcantarilla et al., 2013) Alcantarilla, P., Nuevo, J. & Bartoli, A. Fast explicit diffusion for accelerated features in nonlinear scale spaces. in Procedings of the British Machine Vision Conference 13.1–13.11 (British Machine Vision Assoc., 2013).
(Ronneberger et al., 2015) Ronneberger, O., Fischer, P., & Brox, T. (2015). U-Net: Convolutional networks for biomedical image segmentation. arXiv.
(Klein et al., 2010) Klein, S., Staring, M., Murphy, K., Viergever, M. A., & Pluim, J. P. W. (2010). Elastix: A toolbox for intensity-based medical image registration. IEEE Transactions on Medical Imaging, 29(1), 196–205.
(Shamonin et al., 2014) Shamonin, D. P., Bron, E. E., Lelieveldt, B. P. F., Smits, M., Klein, S., & Staring, M. (2014). Fast parallel image registration on CPU and GPU for diagnostic classification of Alzheimer's disease. Frontiers in Neuroinformatics, 7, 50.
(Filiot et al., 2025) Filiot, A., Dop, N., Tchita, O., Riou, A., Peeters, T., Valter, D., Scalbert, M., Saillard, C., Robin, G., & Olivier, A. (2025). Distilling foundation models for robust and efficient models in digital pathology. arXiv. https://arxiv.org/abs/2501.16239
- Downloads last month
- 390