Dataset Viewer
script
stringlengths 113
767k
|
|---|
import numpy as np
import pandas as pd
from pandas_profiling import ProfileReport
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# # 1. Load Data & Check Information
df_net = pd.read_csv("../input/netflix-shows/netflix_titles.csv")
# dfqwefdsafqwe
df_net.head()
ProfileReport(df_net)
|
import pandas as pd
import numpy as np
import os
import matplotlib.pyplot as plt
from keras import models
from keras.utils import to_categorical, np_utils
from tensorflow import convert_to_tensor
from tensorflow.image import grayscale_to_rgb
from tensorflow.data import Dataset
from tensorflow.keras.layers import Flatten, Dense, GlobalAvgPool2D, GlobalMaxPool2D
from tensorflow.keras.callbacks import Callback, EarlyStopping, ReduceLROnPlateau
from tensorflow.keras import optimizers
from tensorflow.keras.utils import plot_model
# import tensorflow as tf
# tf.__version__
# Define the input path and show all files
path = "/kaggle/input/challenges-in-representation-learning-facial-expression-recognition-challenge/"
os.listdir(path)
# Load the image data with labels.
data = pd.read_csv(path + "icml_face_data.csv")
data.head()
# Overview
data[" Usage"].value_counts()
emotions = {
0: "Angry",
1: "Disgust",
2: "Fear",
3: "Happy",
4: "Sad",
5: "Surprise",
6: "Neutral",
}
def prepare_data(data):
"""Prepare data for modeling
input: data frame with labels und pixel data
output: image and label array"""
image_array = np.zeros(shape=(len(data), 48, 48))
image_label = np.array(list(map(int, data["emotion"])))
for i, row in enumerate(data.index):
image = np.fromstring(data.loc[row, " pixels"], dtype=int, sep=" ")
image = np.reshape(image, (48, 48))
image_array[i] = image
return image_array, image_label
# Define training, validation and test data:
train_image_array, train_image_label = prepare_data(data[data[" Usage"] == "Training"])
val_image_array, val_image_label = prepare_data(data[data[" Usage"] == "PrivateTest"])
test_image_array, test_image_label = prepare_data(data[data[" Usage"] == "PublicTest"])
# Reshape and scale the images:
train_images = train_image_array.reshape((train_image_array.shape[0], 48, 48, 1))
train_images = train_images.astype("float32") / 255
val_images = val_image_array.reshape((val_image_array.shape[0], 48, 48, 1))
val_images = val_images.astype("float32") / 255
test_images = test_image_array.reshape((test_image_array.shape[0], 48, 48, 1))
test_images = test_images.astype("float32") / 255
# As the pretrained model expects rgb images, we convert our grayscale images with a single channel to pseudo-rgb images with 3 channels
train_images_rgb = grayscale_to_rgb(convert_to_tensor(train_images))
val_images_rgb = grayscale_to_rgb(convert_to_tensor(val_images))
test_images_rgb = grayscale_to_rgb(convert_to_tensor(test_images))
# Data Augmentation using ImageDataGenerator
# sources:
# https://www.tensorflow.org/api_docs/python/tf/keras/preprocessing/image/ImageDataGenerator
# https://pyimagesearch.com/2019/07/08/keras-imagedatagenerator-and-data-augmentation/
from tensorflow.keras.preprocessing.image import ImageDataGenerator
train_rgb_datagen = ImageDataGenerator(
rotation_range=0.15,
width_shift_range=0.15,
height_shift_range=0.15,
shear_range=0.15,
zoom_range=0.15,
horizontal_flip=True,
zca_whitening=False,
)
train_rgb_datagen.fit(train_images_rgb)
# Encoding of the target value:
train_labels = to_categorical(train_image_label)
val_labels = to_categorical(val_image_label)
test_labels = to_categorical(test_image_label)
def plot_examples(label=0):
fig, axs = plt.subplots(1, 5, figsize=(25, 12))
fig.subplots_adjust(hspace=0.2, wspace=0.2)
axs = axs.ravel()
for i in range(5):
idx = data[data["emotion"] == label].index[i]
axs[i].imshow(train_images[idx][:, :, 0], cmap="gray")
axs[i].set_title(emotions[train_labels[idx].argmax()])
axs[i].set_xticklabels([])
axs[i].set_yticklabels([])
plot_examples(label=0)
plot_examples(label=1)
plot_examples(label=2)
plot_examples(label=3)
plot_examples(label=4)
plot_examples(label=5)
plot_examples(label=6)
# In case we may want to save some examples:
from PIL import Image
def save_all_emotions(channels=1, imgno=0):
for i in range(7):
idx = data[data["emotion"] == i].index[imgno]
emotion = emotions[train_labels[idx].argmax()]
img = train_images[idx]
if channels == 1:
img = img.squeeze()
else:
img = grayscale_to_rgb(
convert_to_tensor(img)
).numpy() # convert to tensor, then to 3ch, back to numpy
img_shape = img.shape
# print(f'img shape: {img_shape[0]},{img_shape[1]}, type: {type(img)}') #(48,48)
img = img * 255
img = img.astype(np.uint8)
suf = "_%d_%d_%d" % (img_shape[0], img_shape[1], channels)
os.makedirs("examples" + suf, exist_ok=True)
fname = os.path.join("examples" + suf, emotion + suf + ".png")
Image.fromarray(img).save(fname)
print(f"saved: {fname}")
save_all_emotions(channels=3, imgno=0)
def plot_compare_distributions(array1, array2, title1="", title2=""):
df_array1 = pd.DataFrame()
df_array2 = pd.DataFrame()
df_array1["emotion"] = array1.argmax(axis=1)
df_array2["emotion"] = array2.argmax(axis=1)
fig, axs = plt.subplots(1, 2, figsize=(12, 6), sharey=False)
x = emotions.values()
y = df_array1["emotion"].value_counts()
keys_missed = list(set(emotions.keys()).difference(set(y.keys())))
for key_missed in keys_missed:
y[key_missed] = 0
axs[0].bar(x, y.sort_index(), color="orange")
axs[0].set_title(title1)
axs[0].grid()
y = df_array2["emotion"].value_counts()
keys_missed = list(set(emotions.keys()).difference(set(y.keys())))
for key_missed in keys_missed:
y[key_missed] = 0
axs[1].bar(x, y.sort_index())
axs[1].set_title(title2)
axs[1].grid()
plt.show()
plot_compare_distributions(
train_labels, val_labels, title1="train labels", title2="val labels"
)
# Calculate the class weights of the label distribution:
class_weight = dict(
zip(
range(0, 7),
(
(
(
data[data[" Usage"] == "Training"]["emotion"].value_counts()
).sort_index()
)
/ len(data[data[" Usage"] == "Training"]["emotion"])
).tolist(),
)
)
class_weight
# ## General defintions and helper functions
# Define callbacks
early_stopping = EarlyStopping(
monitor="val_accuracy",
min_delta=0.00008,
patience=11,
verbose=1,
restore_best_weights=True,
)
lr_scheduler = ReduceLROnPlateau(
monitor="val_accuracy",
min_delta=0.0001,
factor=0.25,
patience=4,
min_lr=1e-7,
verbose=1,
)
callbacks = [
early_stopping,
lr_scheduler,
]
# General shape parameters
IMG_SIZE = 48
NUM_CLASSES = 7
BATCH_SIZE = 64
# A plotting function to visualize training progress
def render_history(history, suf=""):
fig, (ax1, ax2) = plt.subplots(1, 2)
plt.subplots_adjust(left=0.1, bottom=0.1, right=0.95, top=0.9, wspace=0.4)
ax1.set_title("Losses")
ax1.plot(history.history["loss"], label="loss")
ax1.plot(history.history["val_loss"], label="val_loss")
ax1.set_xlabel("epochs")
ax1.set_ylabel("value of the loss function")
ax1.legend()
ax2.set_title("Accuracies")
ax2.plot(history.history["accuracy"], label="accuracy")
ax2.plot(history.history["val_accuracy"], label="val_accuracy")
ax2.set_xlabel("epochs")
ax2.set_ylabel("value of accuracy")
ax2.legend()
plt.show()
suf = "" if suf == "" else "_" + suf
fig.savefig("loss_and_acc" + suf + ".png")
# ## Model construction
from tensorflow.keras.applications import MobileNet
from tensorflow.keras.models import Model
# By specifying the include_top=False argument, we load a network that
# doesn't include the classification layers at the top, which is ideal for feature extraction.
base_net = MobileNet(
input_shape=(IMG_SIZE, IMG_SIZE, 3), include_top=False, weights="imagenet"
)
# plot_model(base_net, show_shapes=True, show_layer_names=True, expand_nested=True, dpi=50, to_file='mobilenet_full.png')
# For these small images, mobilenet is a very large model. Observing that there is nothing left to convolve further, we take the model only until the 12.block
base_model = Model(
inputs=base_net.input,
outputs=base_net.get_layer("conv_pw_12_relu").output,
name="mobilenet_trunc",
)
# this is the same as:
# base_model = Model(inputs = base_net.input,outputs = base_net.layers[-7].output)
# plot_model(base_model, show_shapes=True, show_layer_names=True, expand_nested=True, dpi=50, to_file='mobilenet_truncated.png')
# from: https://www.tensorflow.org/tutorials/images/transfer_learning
from tensorflow.keras import Sequential, layers
from tensorflow.keras import Input, Model
# from tensor
# base_model.trainable = False
# This model expects pixel values in [-1, 1], but at this point, the pixel values in your images are in [0, 255].
# To rescale them, use the preprocessing method included with the model.
# preprocess_input = tf.keras.applications.mobilenet_v2.preprocess_input
# Add a classification head: To generate predictions from the block of features,
# average over the spatial 2x2 spatial locations, using a tf.keras.layers.GlobalAveragePooling2D layer
# to convert the features to a single 1280-element vector per image.
global_average_layer = GlobalAvgPool2D()
# feature_batch_average = global_average_layer(feature_batch)
# print(feature_batch_average.shape)
# Apply a tf.keras.layers.Dense layer to convert these features into a single prediction per image.
# You don't need an activation function here because this prediction will be treated as a logit,
# or a raw prediction value. Positive numbers predict class 1, negative numbers predict class 0.
prediction_layer = Dense(NUM_CLASSES, activation="softmax", name="pred")
# prediction_batch = prediction_layer(feature_batch_average)
# print(prediction_batch.shape)
# Build a model by chaining together the data augmentation, rescaling, base_model and feature extractor layers
# using the Keras Functional API. As previously mentioned, use training=False as our model contains a BatchNormalization layer.
inputs_raw = Input(shape=(IMG_SIZE, IMG_SIZE, 3))
# inputs_pp = preprocess_input(inputs_aug)
# x = base_model(inputs_pp, training=False)
x = base_model(inputs_raw, training=False)
x = global_average_layer(x)
# x = tf.keras.layers.Dropout(0.2)(x)
outputs = prediction_layer(x)
model = Model(inputs=inputs_raw, outputs=outputs)
model.summary()
plot_model(
model,
show_shapes=True,
show_layer_names=True,
expand_nested=True,
dpi=50,
to_file="MobileNet12blocks_structure.png",
)
# Train the classification head:
# base_model.trainable = True #if we included the model layers, but not the model itself, this doesn't have any effect
for layer in base_model.layers[:]:
layer.trainable = False
# for layer in base_model.layers[81:]:
# layer.trainable = True
optims = {
"sgd": optimizers.SGD(lr=0.1, momentum=0.9, decay=0.01),
"adam": optimizers.Adam(0.01),
"nadam": optimizers.Nadam(
learning_rate=0.1, beta_1=0.9, beta_2=0.999, epsilon=1e-07
),
}
model.compile(
loss="categorical_crossentropy", optimizer=optims["adam"], metrics=["accuracy"]
)
model.summary()
initial_epochs = 5
# total_epochs = initial_epochs + 5
history = model.fit_generator(
train_rgb_datagen.flow(train_images_rgb, train_labels, batch_size=BATCH_SIZE),
validation_data=(val_images_rgb, val_labels),
class_weight=class_weight,
steps_per_epoch=len(train_images) / BATCH_SIZE,
# initial_epoch = history.epoch[-1],
# epochs = total_epochs,
epochs=initial_epochs,
callbacks=callbacks,
use_multiprocessing=True,
)
# ### Fine-tuning
iterative_finetuning = False
# #### First iteration: partial fine-tuning of the base_model
if iterative_finetuning:
# fine-tune the top layers (blocks 7-12):
# Let's take a look to see how many layers are in the base model
print("Number of layers in the base model: ", len(base_model.layers))
# base_model.trainable = True #if we included the model layers, but not the model itself, this doesn't have any effect
for layer in base_model.layers:
layer.trainable = False
for layer in base_model.layers[-37:]: # blocks 7-12
layer.trainable = True
optims = {
"sgd": optimizers.SGD(lr=0.01, momentum=0.9, decay=0.01),
"adam": optimizers.Adam(0.001),
"nadam": optimizers.Nadam(
learning_rate=0.01, beta_1=0.9, beta_2=0.999, epsilon=1e-07
),
}
model.compile(
loss="categorical_crossentropy", optimizer=optims["adam"], metrics=["accuracy"]
)
model.summary()
if iterative_finetuning:
fine_tune_epochs = 40
total_epochs = history.epoch[-1] + fine_tune_epochs
history = model.fit_generator(
train_rgb_datagen.flow(train_images_rgb, train_labels, batch_size=BATCH_SIZE),
validation_data=(val_images_rgb, val_labels),
class_weight=class_weight,
steps_per_epoch=len(train_images) / BATCH_SIZE,
initial_epoch=history.epoch[-1],
epochs=total_epochs,
callbacks=callbacks,
use_multiprocessing=True,
)
if iterative_finetuning:
test_loss, test_acc = model.evaluate(test_images_rgb, test_labels) # , test_labels
print("test caccuracy:", test_acc)
if iterative_finetuning:
render_history(history, "mobilenet12blocks_wdgenaug_finetuning1")
# #### Second Iteration (or the main iteration, if iterative_finetuning was set to False): fine-tuning of the entire base_model
if iterative_finetuning:
ftsuf = "ft_2"
else:
ftsuf = "ft_atonce"
# fine-tune all layers
# Let's take a look to see how many layers are in the base model
print("Number of layers in the base model: ", len(base_model.layers))
# base_model.trainable = True #if we included the model layers, but not the model itself, this doesn't have any effect
for layer in base_model.layers:
layer.trainable = False
for layer in base_model.layers[:]:
layer.trainable = True
optims = {
"sgd": optimizers.SGD(lr=0.01, momentum=0.9, decay=0.01),
"adam": optimizers.Adam(0.0001),
"nadam": optimizers.Nadam(
learning_rate=0.01, beta_1=0.9, beta_2=0.999, epsilon=1e-07
),
}
model.compile(
loss="categorical_crossentropy", optimizer=optims["adam"], metrics=["accuracy"]
)
model.summary()
fine_tune_epochs = 100
total_epochs = history.epoch[-1] + fine_tune_epochs
history = model.fit_generator(
train_rgb_datagen.flow(train_images_rgb, train_labels, batch_size=BATCH_SIZE),
validation_data=(val_images_rgb, val_labels),
class_weight=class_weight,
steps_per_epoch=len(train_images) / BATCH_SIZE,
initial_epoch=history.epoch[-1],
epochs=total_epochs,
callbacks=callbacks,
use_multiprocessing=True,
)
test_loss, test_acc = model.evaluate(test_images_rgb, test_labels) # , test_labels
print("test caccuracy:", test_acc)
render_history(history, "mobilenet12blocks_wdgenaug_" + ftsuf)
pred_test_labels = model.predict(test_images_rgb)
model_yaml = model.to_yaml()
with open(
"MobileNet12blocks_wdgenaug_onrawdata_valacc_" + ftsuf + ".yaml", "w"
) as yaml_file:
yaml_file.write(model_yaml)
model.save("MobileNet12blocks_wdgenaug_onrawdata_valacc_" + ftsuf + ".h5")
# ### Analyze the predictions made for the test data
def plot_image_and_emotion(
test_image_array, test_image_label, pred_test_labels, image_number
):
"""Function to plot the image and compare the prediction results with the label"""
fig, axs = plt.subplots(1, 2, figsize=(12, 6), sharey=False)
bar_label = emotions.values()
axs[0].imshow(test_image_array[image_number], "gray")
axs[0].set_title(emotions[test_image_label[image_number]])
axs[1].bar(bar_label, pred_test_labels[image_number], color="orange", alpha=0.7)
axs[1].grid()
plt.show()
import ipywidgets as widgets
@widgets.interact
def f(x=106):
# print(x)
plot_image_and_emotion(test_image_array, test_image_label, pred_test_labels, x)
# ### Make inference for a single image from scratch:
def predict_emotion_of_image(
test_image_array, test_image_label, pred_test_labels, image_number
):
input_arr = test_image_array[image_number] / 255
input_arr = input_arr.reshape((48, 48, 1))
input_arr_rgb = grayscale_to_rgb(convert_to_tensor(input_arr))
predictions = model.predict(np.array([input_arr_rgb]))
predictions_f = [
"%s:%5.2f" % (emotions[i], p * 100) for i, p in enumerate(predictions[0])
]
label = emotions[test_image_label[image_number]]
return f"Label: {label}\nPredictions: {predictions_f}"
import ipywidgets as widgets
@widgets.interact
def f(x=106):
result = predict_emotion_of_image(
test_image_array, test_image_label, pred_test_labels, x
)
print(result)
# ## Compare the distribution of labels and predicted labels
def plot_compare_distributions(array1, array2, title1="", title2=""):
df_array1 = pd.DataFrame()
df_array2 = pd.DataFrame()
df_array1["emotion"] = array1.argmax(axis=1)
df_array2["emotion"] = array2.argmax(axis=1)
fig, axs = plt.subplots(1, 2, figsize=(12, 6), sharey=False)
x = emotions.values()
y = df_array1["emotion"].value_counts()
keys_missed = list(set(emotions.keys()).difference(set(y.keys())))
for key_missed in keys_missed:
y[key_missed] = 0
axs[0].bar(x, y.sort_index(), color="orange")
axs[0].set_title(title1)
axs[0].grid()
y = df_array2["emotion"].value_counts()
keys_missed = list(set(emotions.keys()).difference(set(y.keys())))
for key_missed in keys_missed:
y[key_missed] = 0
axs[1].bar(x, y.sort_index())
axs[1].set_title(title2)
axs[1].grid()
plt.show()
plot_compare_distributions(
test_labels, pred_test_labels, title1="test labels", title2="predict labels"
)
df_compare = pd.DataFrame()
df_compare["real"] = test_labels.argmax(axis=1)
df_compare["pred"] = pred_test_labels.argmax(axis=1)
df_compare["wrong"] = np.where(df_compare["real"] != df_compare["pred"], 1, 0)
from sklearn.metrics import confusion_matrix
from mlxtend.plotting import plot_confusion_matrix
conf_mat = confusion_matrix(test_labels.argmax(axis=1), pred_test_labels.argmax(axis=1))
fig, ax = plot_confusion_matrix(
conf_mat=conf_mat,
show_normed=True,
show_absolute=False,
class_names=emotions.values(),
figsize=(8, 8),
)
fig.show()
|
# #### EEMT 5400 IT for E-Commerce Applications
# ##### HW4 Max score: (1+1+1)+(1+1+2+2)+(1+2)+2
# You will use two different datasets in this homework and you can find their csv files in the below hyperlinks.
# 1. Car Seat:
# https://raw.githubusercontent.com/selva86/datasets/master/Carseats.csv
# 2. Bank Personal Loan:
# https://raw.githubusercontent.com/ChaithrikaRao/DataChime/master/Bank_Personal_Loan_Modelling.csv
# #### Q1.
# a) Perform PCA for both datasets. Create the scree plots (eigenvalues).
# b) Suggest the optimum number of compenents for each dataset with explanation.
# c) Save the PCAs as carseat_pca and ploan_pca respectively.
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
from sklearn.decomposition import PCA
from sklearn.preprocessing import StandardScaler
carseat_df = pd.read_csv(
"https://raw.githubusercontent.com/selva86/datasets/master/Carseats.csv"
)
ploan_df = pd.read_csv(
"https://raw.githubusercontent.com/ChaithrikaRao/DataChime/master/Bank_Personal_Loan_Modelling.csv"
)
scaler = StandardScaler()
numeric_carseat = carseat_df.select_dtypes(include=[np.number])
carseat_std = scaler.fit_transform(numeric_carseat)
numeric_ploan = ploan_df.select_dtypes(include=[np.number])
ploan_std = scaler.fit_transform(numeric_ploan)
pca_carseat = PCA()
pca_ploan = PCA()
carseat_pca_result = pca_carseat.fit(carseat_std)
ploan_pca_result = pca_ploan.fit(ploan_std)
def scree_plot(pca_result, title):
plt.figure()
plt.plot(np.cumsum(pca_result.explained_variance_ratio_))
plt.xlabel("Number of Components")
plt.ylabel("Cumulative Explained Variance")
plt.title(title)
plt.show()
scree_plot(carseat_pca_result, "Car Seat Dataset")
scree_plot(ploan_pca_result, "Bank Personal Loan Dataset")
# (b)The optimal number of components can be determined by looking at the point where the cumulative explained variance "elbows" or starts to level off.
# For the Car Seat dataset, it appears to be around 3 components.
# For the Bank Personal Loan dataset, it appears to be around 4 components.
carseat_pca = PCA(n_components=3)
ploan_pca = PCA(n_components=4)
carseat_pca_result = carseat_pca.fit_transform(carseat_std)
ploan_pca_result = ploan_pca.fit_transform(ploan_std)
# #### Q2. (Car Seat Dataset)
# a) Convert the non-numeric variables to numeric by using get_dummies() method in pandas. Use it in this question.
# b) Use the scikit learn variance filter to reduce the dimension of the dataset. Try different threshold and suggest the best one.
# c) Some columns may have high correlation. For each set of highly correlated variables, keep one variable only and remove the rest of highly correlated columns. (Tips: You can find the correlations among columns by using .corr() method of pandas dataframe. Reference: https://pandas.pydata.org/docs/reference/api/pandas.DataFrame.corr.html)
# d) Perform linear regression to predict the Sales with datasets from part b and part c respectively and compare the result
#
carseat_dummies = pd.get_dummies(carseat_df, drop_first=True)
from sklearn.feature_selection import VarianceThreshold
def filter_by_variance(data, threshold):
var_filter = VarianceThreshold(threshold=threshold)
return pd.DataFrame(
var_filter.fit_transform(data), columns=data.columns[var_filter.get_support()]
)
carseat_filtered_001 = filter_by_variance(carseat_dummies, 0.01)
carseat_filtered_01 = filter_by_variance(carseat_dummies, 0.1)
carseat_filtered_1 = filter_by_variance(carseat_dummies, 1)
print(f"0.01 threshold: {carseat_filtered_001.shape[1]} columns")
print(f"0.1 threshold: {carseat_filtered_01.shape[1]} columns")
print(f"1 threshold: {carseat_filtered_1.shape[1]} columns")
def remove_high_corr(data, threshold):
corr_matrix = data.corr().abs()
upper = corr_matrix.where(np.triu(np.ones(corr_matrix.shape), k=1).astype(np.bool))
to_drop = [column for column in upper.columns if any(upper[column] > threshold)]
return data.drop(columns=to_drop)
carseat_no_high_corr = remove_high_corr(carseat_filtered_01, 0.8)
def linear_regression_score(X, y):
X_train, X_test, y_train, y_test = train_test_split(
X, y, test_size=0.3, random_state=42
)
lr = LinearRegression()
lr.fit(X_train, y_train)
y_pred = lr.predict(X_test)
return r2_score(y_test, y_pred)
X_filtered = carseat_filtered_01.drop(columns=["Sales"])
y_filtered = carseat_filtered_01["Sales"]
X_no_high_corr = carseat_no_high_corr.drop(columns=["Sales"])
y_no_high_corr = carseat_no_high_corr["Sales"]
filtered_score = linear_regression_score(X_filtered, y_filtered)
no_high_corr_score = linear_regression_score(X_no_high_corr, y_no_high_corr)
print(f"Filtered dataset R-squared: {filtered_score}")
print(f"No high correlation dataset R-squared: {no_high_corr_score}")
# #### Q3. (Bank Personal Loan Dataset)
# a) Find the variable which has the highest correlations with CCAvg
# b) Perform polynomial regression to predict CCAvg with the variable identified in part a.
# ##### Tips:
# step 1 - convert the dataset to polynomial using PolynomialFeatures from scikit learn (https://scikit-learn.org/stable/modules/generated/sklearn.preprocessing.PolynomialFeatures.html)
# step 2 - Perform linear regression using scikit learn
ploan_df = pd.read_csv(
"https://raw.githubusercontent.com/ChaithrikaRao/DataChime/master/Bank_Personal_Loan_Modelling.csv"
)
correlations = ploan_df.corr().abs()
highest_corr = correlations["CCAvg"].sort_values(ascending=False).index[1]
print(f"The variable with the highest correlation with CCAvg is {highest_corr}")
X = ploan_df[[highest_corr]]
y = ploan_df["CCAvg"]
poly = PolynomialFeatures(degree=2)
X_poly = poly.fit_transform(X)
X_train, X_test, y_train, y_test = train_test_split(
X_poly, y, test_size=0.3, random_state=42
)
lr = LinearRegression()
lr.fit(X_train, y_train)
y_pred = lr.predict(X_test)
poly_r_squared = r2_score(y_test, y_pred)
print(f"Polynomial regression R-squared: {poly_r_squared}")
# #### Q4. (Bank Personal Loan Dataset)
# Perform linear regression with all variables in the dataset and compare the result with the model in question 3 using R-Squared value.
X_all = ploan_df.drop(columns=["ID", "CCAvg"])
y_all = ploan_df["CCAvg"]
X_train, X_test, y_train, y_test = train_test_split(
X_all, y_all, test_size=0.3, random_state=42
)
lr_all = LinearRegression()
lr_all.fit(X_train, y_train)
y_pred_all = lr_all.predict(X_test)
all_r_squared = r2_score(y_test, y_pred_all)
print(f"All variables linear regression R-squared: {all_r_squared}")
print(f"Polynomial regression R-squared: {poly_r_squared}")
print(f"All variables linear regression R-squared: {all_r_squared}")
|
# 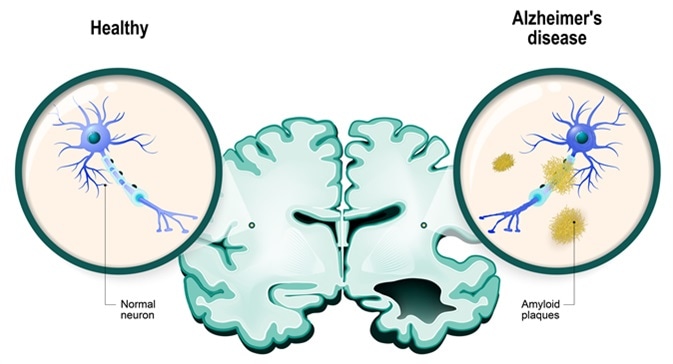
# **Alzheimer's disease** is the most common type of dementia. It is a progressive disease beginning with mild memory loss and possibly leading to loss of the ability to carry on a conversation and respond to the environment. Alzheimer's disease involves parts of the brain that control thought, memory, and language.
# # **Importing libraries**
import pandas as pd
import numpy as np
import os
import matplotlib.pyplot as plt
import warnings
from tensorflow.keras.applications.vgg19 import preprocess_input
from tensorflow.keras.preprocessing import image, image_dataset_from_directory
from tensorflow.keras.preprocessing.image import ImageDataGenerator as IDG
from imblearn.over_sampling import SMOTE
from tensorflow.keras.models import Sequential
from tensorflow import keras
import tensorflow
import tensorflow as tf
from tensorflow.keras.models import Model
from tensorflow.keras.applications.vgg16 import VGG16
from tensorflow.keras.layers import Input, Lambda, Dense, Flatten, Dropout
from tensorflow.keras.models import Model
from sklearn.model_selection import train_test_split
import seaborn as sns
import pathlib
from tensorflow.keras.utils import plot_model
from keras.callbacks import ModelCheckpoint, ReduceLROnPlateau, EarlyStopping
from sklearn.metrics import classification_report, confusion_matrix
# # **Identify dataset**
train_ds = tf.keras.preprocessing.image_dataset_from_directory(
"/kaggle/input/adni-extracted-axial/Axial",
validation_split=0.2,
subset="training",
seed=1337,
image_size=[180, 180],
batch_size=16,
)
val_ds = tf.keras.preprocessing.image_dataset_from_directory(
"/kaggle/input/adni-extracted-axial/Axial",
validation_split=0.2,
subset="validation",
seed=1337,
image_size=[180, 180],
batch_size=16,
)
# number and names of Classes
classnames = train_ds.class_names
len(classnames), train_ds.class_names
# # **Data Visualization**
plt.figure(figsize=(10, 10))
for images, labels in train_ds.take(1):
for i in range(9):
ax = plt.subplot(3, 3, i + 1)
plt.imshow(images[i].numpy().astype("uint8"))
plt.title(train_ds.class_names[labels[i]])
plt.axis("off")
# Number of images in each class
NUM_IMAGES = []
for label in classnames:
dir_name = "/kaggle/input/adni-extracted-axial/Axial" + "/" + label
NUM_IMAGES.append(len([name for name in os.listdir(dir_name)]))
NUM_IMAGES, classnames
# Rename class names
class_names = ["Alzheimer Disease", "Cognitively Impaired", "Cognitively Normal"]
train_ds.class_names = class_names
val_ds.class_names = class_names
NUM_CLASSES = len(class_names)
NUM_CLASSES
# Before Oversampling
# Visualization of each class with pie chart
import matplotlib.pyplot as plt
fig1, ax1 = plt.subplots()
ax1.pie(NUM_IMAGES, autopct="%1.1f%%", labels=train_ds.class_names)
plt.legend(title="Three Classes:", bbox_to_anchor=(0.75, 1.15))
# Performing Image Augmentation to have more data samples
IMG_SIZE = 180
IMAGE_SIZE = [180, 180]
DIM = (IMG_SIZE, IMG_SIZE)
ZOOM = [0.99, 1.01]
BRIGHT_RANGE = [0.8, 1.2]
HORZ_FLIP = True
FILL_MODE = "constant"
DATA_FORMAT = "channels_last"
WORK_DIR = "/kaggle/input/adni-extracted-axial/Axial"
work_dr = IDG(
rescale=1.0 / 255,
brightness_range=BRIGHT_RANGE,
zoom_range=ZOOM,
data_format=DATA_FORMAT,
fill_mode=FILL_MODE,
horizontal_flip=HORZ_FLIP,
)
train_data_gen = work_dr.flow_from_directory(
directory=WORK_DIR, target_size=DIM, batch_size=8000, shuffle=False
)
train_data, train_labels = train_data_gen.next()
# # **Oversampling technique**
# 
# Shape of data before oversampling
print(train_data.shape, train_labels.shape)
# Performing over-sampling of the data, since the classes are imbalanced
# After oversampling using SMOTE
sm = SMOTE(random_state=42)
train_data, train_labels = sm.fit_resample(
train_data.reshape(-1, IMG_SIZE * IMG_SIZE * 3), train_labels
)
train_data = train_data.reshape(-1, IMG_SIZE, IMG_SIZE, 3)
print(train_data.shape, train_labels.shape)
# Show pie plot for dataset (after oversampling)
# Visualization of each class with pie chart
images_after = [2590, 2590, 2590]
import matplotlib.pyplot as plt
fig1, ax1 = plt.subplots()
ax1.pie(images_after, autopct="%1.1f%%", labels=train_ds.class_names)
plt.legend(title="Three Classes:", bbox_to_anchor=(0.75, 1.15))
# # **Spliting data**
train_data, test_data, train_labels, test_labels = train_test_split(
train_data, train_labels, test_size=0.2, random_state=42
)
train_data, val_data, train_labels, val_labels = train_test_split(
train_data, train_labels, test_size=0.2, random_state=42
)
# # **Building the model**
# -------VGG16--------
vgg = VGG16(input_shape=(180, 180, 3), weights="imagenet", include_top=False)
for layer in vgg.layers:
layer.trainable = False
x = Flatten()(vgg.output)
prediction = Dense(3, activation="softmax")(x)
modelvgg = Model(inputs=vgg.input, outputs=prediction)
# Plotting layers as an image
plot_model(modelvgg, to_file="alzahimer.png", show_shapes=True)
# Optimizing model
modelvgg.compile(
optimizer="adam", loss="categorical_crossentropy", metrics=["accuracy"]
)
# Callbacks
checkpoint = ModelCheckpoint(
filepath="best_weights.hdf5", save_best_only=True, save_weights_only=True
)
lr_reduce = ReduceLROnPlateau(
monitor="val_loss", factor=0.3, patience=2, verbose=2, mode="max"
)
early_stop = EarlyStopping(monitor="val_loss", min_delta=0.1, patience=1, mode="min")
# # **Training model using my data**
# Fitting the model
hist = modelvgg.fit(
train_data,
train_labels,
epochs=10,
validation_data=(val_data, val_labels),
callbacks=[checkpoint, lr_reduce],
)
# Plotting accuracy and loss of the model
fig, ax = plt.subplots(1, 2, figsize=(20, 3))
ax = ax.ravel()
for i, met in enumerate(["accuracy", "loss"]):
ax[i].plot(hist.history[met])
ax[i].plot(hist.history["val_" + met])
ax[i].set_title("Model {}".format(met))
ax[i].set_xlabel("epochs")
ax[i].set_ylabel(met)
ax[i].legend(["train", "val"])
# Evaluation using test data
test_scores = modelvgg.evaluate(test_data, test_labels)
print("Testing Accuracy: %.2f%%" % (test_scores[1] * 100))
pred_labels = modelvgg.predict(test_data)
# # **Confusion matrix**
pred_ls = np.argmax(pred_labels, axis=1)
test_ls = np.argmax(test_labels, axis=1)
conf_arr = confusion_matrix(test_ls, pred_ls)
plt.figure(figsize=(8, 6), dpi=80, facecolor="w", edgecolor="k")
ax = sns.heatmap(
conf_arr,
cmap="Greens",
annot=True,
fmt="d",
xticklabels=classnames,
yticklabels=classnames,
)
plt.title("Alzheimer's Disease Diagnosis")
plt.xlabel("Prediction")
plt.ylabel("Truth")
plt.show(ax)
# # **Classification report**
print(classification_report(test_ls, pred_ls, target_names=classnames))
# # **Save the model for a Mobile app as tflite**
export_dir = "/kaggle/working/"
tf.saved_model.save(modelvgg, export_dir)
tflite_model_name = "alzheimerfinaly.tflite"
# Convert the model.
converter = tf.lite.TFLiteConverter.from_saved_model(export_dir)
converter.optimizations = [tf.lite.Optimize.OPTIMIZE_FOR_SIZE]
tflite_model = converter.convert()
tflite_model_file = pathlib.Path(tflite_model_name)
tflite_model_file.write_bytes(tflite_model)
# # **Save the model for a Web app as hdf5**
tf.keras.models.save_model(modelvgg, "Alzheimer_finaly.hdf5")
|
# # Tracking COVID-19 from New York City wastewater
# **TABLE OF CONTENTS**
# * [1. Introduction](#chapter_1)
# * [2. Data exploration](#chapter_2)
# * [3. Analysis](#chapter_3)
# * [4. Baseline model](#chapter_4)
# ## 1. Introduction
# The **New York City OpenData Project** (*link:* __[project home page](https://opendata.cityofnewyork.us)__) has hundreds of open, New York-related datasets available for everyone to use. On the website, all datasets are labeled by different city functions (business, government, education etc.).
# While browsing through different subcategories, I came across data by the Department of Environmental Protection (DEP). One dataset they had made available in public concerned the SARS-CoV-2 gene level concentrations measured in NYC wastewater (*link:* __[dataset page](https://data.cityofnewyork.us/Health/SARS-CoV-2-concentrations-measured-in-NYC-Wastewat/f7dc-2q9f)__). As one can guess, SARS-CoV-2 is the virus causing COVID-19.
# Since I had earlier used the NYC data on COVID (*link:* __[dataset page](https://data.cityofnewyork.us/Health/COVID-19-Daily-Counts-of-Cases-Hospitalizations-an/rc75-m7u3)__) in several notebooks since the pandemic, I decided to create a new notebook combining these two datasets.
# This notebook is a brief exploratory analysis on the relationship between the COVID-causing virus concentrations in NYC wastewater and actual COVID-19 cases detected in New York. Are these two related, and if so, how? Let's find out.
# *All data sources are read directly from the OpenData Project website, so potential errors are caused by temporary issues (update etc.) in the website data online availability.*
# **April 13th, 2023
# Jari Peltola**
# ******
# ## 2. Data Exploration
# import modules
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
from sklearn.ensemble import RandomForestRegressor
from sklearn.model_selection import train_test_split
from sklearn.preprocessing import MinMaxScaler
from sklearn.metrics import r2_score, mean_absolute_error, mean_squared_error
from sklearn.model_selection import cross_val_score
# set column and row display
pd.set_option("display.max_columns", None)
pd.set_option("display.max_rows", None)
# disable warnings
pd.options.mode.chained_assignment = None
# wastewater dataset url
url = "https://data.cityofnewyork.us/api/views/f7dc-2q9f/rows.csv?accessType=DOWNLOAD"
# read the data from url
data = pd.read_csv(url, low_memory=False)
# drop rows with NaN values
data = data.dropna()
# reset index
data.reset_index(drop=True, inplace=True)
data.head()
data.shape
# A large metropolis area such as New York City has several wastewater resource recovery facilities (WRRF). Each of these facilities takes care of a specific area in the city's wastewater system (*link:* __[list of NYC wastewater treatment facilities](https://www.nyc.gov/site/dep/water/wastewater-treatment-plants.page)__). The 14 facilities serve city population ranging from less than one hundred thousand (Rockaway) up to one million people (Newtown Creek, Wards Island).
# We start by adding some additional data from the webpage linked above. For each facility, the added data includes receiving waterbody, drainage acres as well as verbal description of the drainage area. This is done to get a better comprehension of where exactly in the city each facility is located. Another possible solution would be to create some kind of map of all WRRF facilities in the area, but since we are actually dealing with water with ever-evolving drainage instead of buildings, this approach would not in my opinion work too well.
# It's good to keep the original data intact, so first we make a list **wrrf_list** consisting of all unique WRRF names, create a new dataframe **df_location** and add the facility names as a column **WRRF Name**.
# create list of all pickup locations
wrrf_list = data["WRRF Name"].unique()
# create new dataframe with empty column
df_location = pd.DataFrame(columns=["WRRF Name"])
# add column values from list
df_location["WRRF Name"] = np.array(wrrf_list)
df_location.head()
df_location["WRRF Name"].unique()
# Next we add three more columns including the additional data.
# list of receiving waterbodies
Receiving_Waterbody = [
"Jamaica Bay",
"Jamaica Bay",
"East River",
"Jamaica Bay",
"Upper East River",
"Jamaica Bay",
"Lower East River",
"Hudson River",
"Upper New York Bay",
"Lower New York Bay",
"Upper East River",
"Upper East River",
"Kill Van Kull",
"Upper East River",
]
# list of drainage acres
Drainage_Acres = [
5907,
15087,
15656,
6259,
16860,
25313,
3200,
6030,
12947,
10779,
16664,
12056,
9665,
15203,
]
# list of drainage areas
Drainage_Area = [
"Eastern section of Brooklyn, near Jamaica Bay",
"South and central Brooklyn",
"South and eastern midtown sections of Manhattan, northeast section of Brooklyn and western section of Queens",
"Rockaway Peninsula",
"Northeast section of Queens",
"Southern section of Queens",
"Northwest section of Brooklyn and Governors Island",
"West side of Manhattan above Bank Street",
"Western section of Brooklyn",
"Southern section of Staten Island",
"Eastern section of the Bronx",
"Western section of the Bronx and upper east side of Manhattan",
"Northern section of Staten Island",
"Northeast section of Queens",
]
# add new columns
df_location["Receiving_Waterbody"] = np.array(Receiving_Waterbody)
df_location["Drainage_Acres"] = np.array(Drainage_Acres)
df_location["Drainage_Area"] = np.array(Drainage_Area)
df_location.head()
# Now we may merge the **df_location** dataframe with our original data. For this we can use the common column **WRRF Name**. For clarity we keep calling also the merged results **data**.
# merge dataframes
data = pd.merge(data, df_location, on="WRRF Name", how="left")
data.dtypes
data.head()
# The date data we have is not in datetime format, so we will take care of that next, along with renaming some columns and dropping others.
# column to datetime
data["Test date"] = pd.to_datetime(data["Test date"])
# rename columns by their index location
data = data.rename(columns={data.columns[1]: "TestDate"})
data = data.rename(columns={data.columns[4]: "SARS_CoV2_Concentration"})
data = data.rename(columns={data.columns[5]: "SARS_CoV2_Concentration_PerCap"})
data = data.rename(columns={data.columns[7]: "Population"})
# drop columns by their index location
data = data.drop(data.columns[[0, 3, 6]], axis=1)
data.head()
# Let's take a look at the COVID concentration when some specific WRRF facilities and their respective waterbodies are concerned. For this example, we choose the **Coney Island** and **Jamaica Bay** facilities and plot the result.
# select facility data to two dataframes
df_coney = data.loc[data["WRRF Name"] == "Coney Island"]
df_jamaica = data.loc[data["WRRF Name"] == "Jamaica Bay"]
# set figure size
plt.figure(figsize=(10, 8))
# set parameters
plt.plot(
df_coney.TestDate,
df_coney.SARS_CoV2_Concentration,
label="Coney Island",
linewidth=3,
)
plt.plot(
df_jamaica.TestDate,
df_jamaica.SARS_CoV2_Concentration,
color="red",
label="Jamaica Bay",
linewidth=3,
)
# add title and axis labels
plt.title(
"COVID virus concentration (Coney Island, Jamaica Bay)", weight="bold", fontsize=16
)
plt.xlabel("Date", weight="bold", fontsize=14)
plt.ylabel("Concentration", weight="bold", fontsize=14)
# add legend
plt.legend()
plt.show()
# We can see that the COVID virus concentration in the two selected wastewater facilites is for the most time pretty similar. The big exception is the New Year period 2021-2022, when Coney Island recorded a significant concentration spike, but in Jamaica Bay the change was much more moderate.
# Next we bring in the New York City COVID-19 dataset and upload it as dataframe **covid_cases**.
# COVID-19 dataset url
url = "https://data.cityofnewyork.us/api/views/rc75-m7u3/rows.csv?accessType=DOWNLOAD"
# read the data from url
covid_cases = pd.read_csv(url, low_memory=False)
# drop rows with NaN values
covid_cases = covid_cases.dropna()
covid_cases.head()
covid_cases.dtypes
# One thing we can see is that the COVID-19 data includes both overall data and more specific figures on the city's five boroughs (the Bronx, Brooklyn, Manhattan, Queens, Staten Island).
# In order to merge the data, we rename and reformat the **date_of_interest** column to fit our existing data, since that's the common column we will use.
# rename column
covid_cases = covid_cases.rename(columns={covid_cases.columns[0]: "TestDate"})
# change format to datetime
covid_cases["TestDate"] = pd.to_datetime(covid_cases["TestDate"])
# merge dataframes
data = pd.merge(data, covid_cases, on="TestDate", how="left")
data.head()
data.shape
# What we don't know is the workload of different facilities when it comes to wastewater treatment. Let's see that next.
# check percentages
wrrf_perc = data["WRRF Name"].value_counts(normalize=True) * 100
wrrf_perc
# The percentages tell us that there is no dominant facility when it comes to wastewater treatment in New York City: it's all one big puzzle with relatively equal pieces.
# Now we are ready to find out more about the potential relationship between COVID virus concentration in wastewater and actual COVID cases.
# ******
# ## 3. Analysis
# First it would be good to know more about boroughs and their different wastewater facilities. Mainly it would be useful to find out if there are differences between wastewater treatment facilities and their measured COVID virus concentration levels when a particular borough is concerned.
# Let's take Brooklyn for example. Based on the verbal descriptions of different wastewater facilities, Brooklyn area is mostly served by the **26th Ward**, **Coney Island** and **Owl's Head** facilities. Next we select those three locations with all necessary columns to dataframe **brooklyn_data**.
# select Brooklyn data
brooklyn_data = data.loc[
data["WRRF Name"].isin(["26th Ward", "Coney Island", "Owls Head"])
]
# select columns
brooklyn_data = brooklyn_data.iloc[:, np.r_[0:8, 29:40]]
brooklyn_data.shape
brooklyn_data.head()
# To access the data more conveniently, we take a step back and create separate dataframes for the three facilities before plotting the result.
# create dataframes
df_26th = brooklyn_data.loc[brooklyn_data["WRRF Name"] == "26th Ward"]
df_coney = brooklyn_data.loc[brooklyn_data["WRRF Name"] == "Coney Island"]
df_owls = brooklyn_data.loc[brooklyn_data["WRRF Name"] == "Owls Head"]
# set figure size
plt.figure(figsize=(10, 8))
# set parameters
plt.plot(df_26th.TestDate, df_26th.SARS_CoV2_Concentration, label="26th", linewidth=3)
plt.plot(
df_coney.TestDate,
df_coney.SARS_CoV2_Concentration,
color="red",
label="Coney Island",
linewidth=3,
)
plt.plot(
df_owls.TestDate,
df_owls.SARS_CoV2_Concentration,
color="green",
label="Owls Head",
linewidth=3,
)
# add title and axis labels
plt.title(
"Virus concentration (26th Ward, Coney Island, Owls Head)",
weight="bold",
fontsize=16,
)
plt.xlabel("Date", weight="bold", fontsize=14)
plt.ylabel("Concentration", weight="bold", fontsize=14)
# add legend
plt.legend()
plt.show()
# There are some differences in concentration intensity, but all in all the figures pretty much follow the same pattern. Also, one must remember that for example the Coney Island facility serves population about twice as large as the Owl's Head facility. Then again, this makes the data from beginning of year 2023 even more intriguing, since Owl's Head had much larger COVID concentration then compared to other two Brooklyn facilities.
# Next we plot the third Brooklyn facility (26th Ward) data and compare it to the 7-day average of hospitalized COVID-19 patients in the area. It is notable that in the plot the concentration is multiplied by 0.01 to make a better visual fit with patient data. This change is made merely for plotting purposes and does not alter the actual values in our data.
plt.figure(figsize=(10, 8))
plt.plot(
df_26th.TestDate,
df_26th.SARS_CoV2_Concentration * 0.01,
label="CoV-2 concentration",
linewidth=3,
)
plt.plot(
df_26th.TestDate,
df_26th.BK_HOSPITALIZED_COUNT_7DAY_AVG,
color="red",
label="Hospitalizations avg",
linewidth=3,
)
# add title and axis labels
plt.title(
"Virus concentration in 26th Ward and COVID hospitalizations",
weight="bold",
fontsize=16,
)
plt.xlabel("Date", weight="bold", fontsize=14)
plt.ylabel("Count", weight="bold", fontsize=14)
# add legend
plt.legend()
# display plot
plt.show()
# The two lines definitely share a similar pattern, which in the end is not that surprising considering they actually describe two different viewpoints to the same phenomenon (COVID-19 pandemic/endemic).
# Next we take a closer look at the first 18 months of the COVID-19 pandemic and narrow our date selection accordingly. Also, we change our COVID-19 measurement to daily hospitalized patients instead of a weekly average.
# mask dataframe
start_date = "2020-01-01"
end_date = "2021-12-31"
# wear a mask
mask = (df_26th["TestDate"] >= start_date) & (df_26th["TestDate"] < end_date)
df_26th_mask = df_26th.loc[mask]
plt.figure(figsize=(10, 8))
plt.plot(
df_26th_mask.TestDate,
df_26th_mask.SARS_CoV2_Concentration * 0.01,
label="CoV-2 concentration",
linewidth=3,
)
plt.plot(
df_26th_mask.TestDate,
df_26th_mask.BK_HOSPITALIZED_COUNT,
color="red",
label="Hospitalizations",
linewidth=3,
)
plt.title(
"26th Ward virus concentration and daily hospitalizations",
weight="bold",
fontsize=16,
)
plt.xlabel("Date", weight="bold", fontsize=14)
plt.ylabel("Count", weight="bold", fontsize=14)
plt.legend()
plt.show()
# Now the similarities are even more clear. Taking a look at the late 2020/early 2021 situation, it seems that **some sort of threshold of daily COVID virus concentration difference in wastewater might actually become a valid predictor for future hospitalizations**, if one omitted the viewpoint of a ML model constructor.
# In this notebook we will not go that far, but next a simple baseline model is created as a sort of first step toward that goal.
# ******
# ## 4. Baseline model
# The regressive baseline model we will produce is all about comparing relative (per capita) COVID virus concentration in wasterwater with different features presented in COVID-19 patient data.
# For our baseline model we will use wastewater data from all five boroughs and their wastewater facilities, meaning we must return to our original **data**.
data.head(2)
data.shape
# As we need only some of the numerical data here, next we select the approriate columns to a new dataframe **df_model**.
# select columns by index
df_model = data.iloc[:, np.r_[3, 8:18]]
df_model.head()
df_model.dtypes
# To ensure better compatibility between different data features, the dataframe is next scaled with MinMaxScaler. This is not a necessary step, but as the COVID virus concentration per capita values are relatively large compared to COVID patient data, by scaling all data we may get a bit better results in actual baseline modeling.
# scale data
scaler = MinMaxScaler()
scaler.fit(df_model)
scaled = scaler.fit_transform(df_model)
scaled_df = pd.DataFrame(scaled, columns=df_model.columns)
scaled_df.head()
# plot scatterplot of COVID-19 case count and virus concentration level
scaled_df.plot(kind="scatter", x="SARS_CoV2_Concentration_PerCap", y="CASE_COUNT")
plt.show()
# Some outliers excluded, the data we have is relatively well concentrated and should therefore fit regressive model pretty well. Just to make sure, we drop potential NaN rows before proceeding.
# drop NaN rows
data = scaled_df.dropna()
# set random seed
np.random.seed(42)
# select features
X = data.drop("SARS_CoV2_Concentration_PerCap", axis=1)
y = data["SARS_CoV2_Concentration_PerCap"]
# separate data into training and validation sets
X_train, X_test, y_train, y_test = train_test_split(
X, y, train_size=0.8, test_size=0.2, random_state=1
)
# define model
model = RandomForestRegressor()
# fit model
model.fit(X_train, y_train)
# Now we can make predictions with the model based on our data.
y_preds = model.predict(X_test)
print("Regression model metrics on test set")
print(f"R2: {r2_score(y_test, y_preds)}")
print(f"MAE: {mean_absolute_error(y_test, y_preds)}")
print(f"MSE: {mean_squared_error(y_test, y_preds)}")
# The worst R2 score is basically minus infinity and the perfect score 1, with zero predicting the mean value every time. Taking this into account, the score of about 0.5 (the exact result changes every time the notebook is run as the online datasets are constantly updated) for a test set score is quite typical when using only a simple baseline fit.
# Also, as seen below, the score on train dataset is significantly better than the one of test dataset. As we had less than 500 datapoints i.e. rows overall for the baseline model to learn from, this is in the end not a big surprise.
print("Train score:")
print(model.score(X_train, y_train))
print("Test score:")
print(model.score(X_test, y_test))
# We can also use the cross-valuation score to run the same baseline model several times and take the average R2 score of the process. In this case we will run the model five times (cv=5).
np.random.seed(42)
# create regressor
rf_reg = RandomForestRegressor(n_estimators=100)
# create five models for cross-valuation
# setting "scoring=None" uses default scoring parameter
# cross_val_score function's default scoring parameter is R2
cv_r2 = cross_val_score(rf_reg, X_train, y_train, cv=5, scoring=None)
# print average R2 score of five baseline regressor models
np.mean(cv_r2)
|
# ## Project 4
# We're going to start with the dataset from Project 1.
# This time the goal is to compare data wrangling runtime by either using **Pandas** or **Polar**.
data_dir = "/kaggle/input/project-4-dataset/data-p1"
sampled = False
path_suffix = "" if not sampled else "_sampled"
from time import time
import pandas as pd
import numpy as np
import polars as pl
from polars import col
def print_time(t):
"""Function that converts time period in seconds into %m:%s:%ms expression.
Args:
t (int): time period in seconds
Returns:
s (string): time period formatted
"""
ms = t * 1000
m, ms = divmod(ms, 60000)
s, ms = divmod(ms, 1000)
return "%dm:%ds:%dms" % (m, s, ms)
# ## Load data
# #### Pandas
start = time()
pandas_data = pd.read_csv(f"{data_dir}/transactions_data{path_suffix}.csv")
print("\nProcessing took {}".format(print_time(time() - start)))
start = time()
pandas_data["date"] = pd.to_datetime(pandas_data["date"])
print("\nProcessing took {}".format(print_time(time() - start)))
## Create sales column
start = time()
pandas_data = (
pandas_data.groupby(
[
pandas_data.date.dt.date,
"id",
"item_id",
"dept_id",
"cat_id",
"store_id",
"state_id",
]
)
.agg("count")
.rename(columns={"date": "sales"})
.reset_index()
.assign(date=lambda df: pd.to_datetime(df.date))
)
print("\nProcessing took {}".format(print_time(time() - start)))
## Convert data types
start = time()
pandas_data = pandas_data.assign(
id=pandas_data.id.astype("category"),
item_id=pandas_data.item_id.astype("category"),
dept_id=pandas_data.dept_id.astype("category"),
cat_id=pandas_data.cat_id.astype("category"),
store_id=pandas_data.store_id.astype("category"),
state_id=pandas_data.state_id.astype("category"),
)
print("\nProcessing took {}".format(print_time(time() - start)))
## Filling with zeros
start = time()
pandas_data = pandas_data.set_index(["date", "id"])
min_date, max_date = (
pandas_data.index.get_level_values("date").min(),
pandas_data.index.get_level_values("date").max(),
)
dates_to_select = pd.date_range(min_date, max_date, freq="1D")
ids = pandas_data.index.get_level_values("id").unique()
index_to_select = pd.MultiIndex.from_product(
[dates_to_select, ids], names=["date", "id"]
)
def fill_category_nans(df, col_name, level_start, level_end):
return np.where(
df[col_name].isna(),
df.index.get_level_values("id")
.str.split("_")
.str[level_start:level_end]
.str.join("_"),
df[col_name],
)
pandas_data = (
pandas_data.reindex(index_to_select)
.fillna({"sales": 0})
.assign(
sales=lambda df: df.sales.astype("int"),
item_id=lambda df: fill_category_nans(df, "item_id", 0, 3),
dept_id=lambda df: fill_category_nans(df, "dept_id", 0, 2),
cat_id=lambda df: fill_category_nans(df, "cat_id", 0, 1),
store_id=lambda df: fill_category_nans(df, "store_id", 3, 5),
state_id=lambda df: fill_category_nans(df, "state_id", 3, 4),
)
.assign(
item_id=lambda df: df.item_id.astype("category"),
dept_id=lambda df: df.dept_id.astype("category"),
cat_id=lambda df: df.cat_id.astype("category"),
store_id=lambda df: df.store_id.astype("category"),
state_id=lambda df: df.state_id.astype("category"),
)
)
print("\nProcessing took {}".format(print_time(time() - start)))
# #### Polars
start = time()
transactions_pl = pl.read_csv(f"{data_dir}/transactions_data{path_suffix}.csv")
print("\nProcessing took {}".format(print_time(time() - start)))
start = time()
transactions_pl = transactions_pl.with_columns(
pl.col("date").str.strptime(pl.Date, fmt="%Y-%m-%d %H:%M:%S", strict=False)
)
print("\nProcessing took {}".format(print_time(time() - start)))
## Create sales column
start = time()
polars_data = (
transactions_pl.lazy()
.with_column(pl.lit(1).alias("sales"))
.groupby(["date", "id", "item_id", "dept_id", "cat_id", "store_id", "state_id"])
.agg(pl.col("sales").sum())
.collect()
)
print("\nProcessing took {}".format(print_time(time() - start)))
## Convert data types
start = time()
polars_data = (
polars_data.lazy()
.with_columns(
[
pl.col("id").cast(pl.Categorical),
pl.col("item_id").cast(pl.Categorical),
pl.col("dept_id").cast(pl.Categorical),
pl.col("cat_id").cast(pl.Categorical),
pl.col("store_id").cast(pl.Categorical),
pl.col("state_id").cast(pl.Categorical),
]
)
.collect()
)
print("\nProcessing took {}".format(print_time(time() - start)))
## Filling with zeros
start = time()
min_date, max_date = (
polars_data.with_columns(pl.col("date")).min()["date"][0],
polars_data.with_columns(pl.col("date")).max()["date"][0],
)
dates_to_select = pl.date_range(min_date, max_date, "1d")
# df with all combinations of daily dates and ids
date_id_df = pl.DataFrame({"date": dates_to_select}).join(
polars_data.select(pl.col("id").unique()), how="cross"
)
# join with original df
polars_data = polars_data.join(date_id_df, on=["date", "id"], how="outer").sort(
"id", "date"
)
# create tmp columns to assemble strings from item_id to fill columns for cells with null values
polars_data = (
polars_data.lazy()
.with_columns(
[
col("id"),
*[
col("id").apply(lambda s, i=i: s.split("_")[i]).alias(col_name)
for i, col_name in enumerate(["1", "2", "3", "4", "5", "6"])
],
]
)
.drop(["item_id", "dept_id", "cat_id", "store_id", "state_id"])
.collect()
)
# concat string components
item_id = polars_data.select(
pl.concat_str(
[
pl.col("1"),
pl.col("2"),
pl.col("3"),
],
separator="_",
).alias("item_id")
)
dept_id = polars_data.select(
pl.concat_str(
[
pl.col("1"),
pl.col("2"),
],
separator="_",
).alias("dept_id")
)
cat_id = polars_data.select(
pl.concat_str(
[
pl.col("1"),
],
separator="_",
).alias("cat_id")
)
store_id = polars_data.select(
pl.concat_str(
[
pl.col("4"),
pl.col("5"),
],
separator="_",
).alias("store_id")
)
state_id = polars_data.select(
pl.concat_str(
[
pl.col("4"),
],
separator="_",
).alias("state_id")
)
# fill sales columns with null values with 0
polars_data = (
polars_data.lazy()
.with_column(
pl.col("sales").fill_null(0),
)
.collect()
)
# recreate other columns with the string components
polars_data = (
pl.concat(
[polars_data, item_id, dept_id, cat_id, store_id, state_id], how="horizontal"
)
.drop(["1", "2", "3", "4", "5", "6"])
.sort("date", "id")
)
print("\nProcessing took {}".format(print_time(time() - start)))
# #### Comparison
polars_data.sort("sales", descending=True).head()
len(polars_data)
pandas_data.reset_index(drop=True).sort_values(by=["sales"], ascending=False).head()
len(pandas_data)
|
# This notebook reveals my solution for __RFM Analysis Task__ offered by Renat Alimbekov.
# This task is part of the __Task Series__ for Data Analysts/Scientists
# __Task Series__ - is a rubric where Alimbekov challenges his followers to solve tasks and share their solutions.
# So here I am :)
# Original solution can be found at - https://alimbekov.com/rfm-python/
# The task is to perform RFM Analysis.
# * __olist_orders_dataset.csv__ and __olist_order_payments_dataset.csv__ should be used
# * order_delivered_carrier_date - should be used in this task
# * Since the dataset is not actual by 2021, thus we should assume that we were asked to perform RFM analysis the day after the last record
# # Importing the modules
import pandas as pd
import numpy as np
import squarify
import matplotlib.pyplot as plt
import seaborn as sns
plt.style.use("ggplot")
import warnings
warnings.filterwarnings("ignore")
# # Loading the data
orders = pd.read_csv("../input/brazilian-ecommerce/olist_orders_dataset.csv")
payments = pd.read_csv("../input/brazilian-ecommerce/olist_order_payments_dataset.csv")
# # Dataframes join
orders["order_delivered_carrier_date"] = pd.to_datetime(
orders["order_delivered_carrier_date"]
) # datetime conversion
payments = payments.set_index("order_id") # preparation before the join
orders = orders.set_index("order_id") # preparation before the join
joined = orders.join(payments) # join on order_id
joined.isna().sum().sort_values(ascending=False)
joined.nunique().sort_values(ascending=False)
# It seems like we have missing values. And unfortunately order_delivered_carrier_date also has missing values. Thus, they should be dropped
last_date = joined["order_delivered_carrier_date"].max() + pd.to_timedelta(1, "D")
RFM = (
joined.dropna(subset=["order_delivered_carrier_date"])
.reset_index()
.groupby("customer_id")
.agg(
Recency=("order_delivered_carrier_date", lambda x: (last_date - x.max()).days),
Frequency=("order_id", "size"),
Monetary=("payment_value", "sum"),
)
)
# Sanity check - do we have NaN values or not?
RFM.isna().sum()
RFM.describe([0.01, 0.05, 0.1, 0.25, 0.5, 0.75, 0.9, 0.95, 0.99]).T
# So, here we can see that we have some outliers in Freqency and Monetary groups. Thus, they should be dropped and be analyzed separately
# # Recency
plt.figure(figsize=(12, 6))
sns.boxplot(x="Recency", data=RFM)
plt.title("Boxplot of Recency")
# # Frequency
RFM["Frequency"].value_counts(normalize=True) * 100
# I guess here we should select only frequency values that are greater than 5, because by doing this we only drop 0.11% of records
RFM["Frequency"].apply(
lambda x: "less or equal to 5" if x <= 5 else "greater than 5"
).value_counts(normalize=True) * 100
RFM = RFM[RFM["Frequency"] <= 5]
# # Monetary
RFM["Monetary"].describe([0.25, 0.5, 0.75, 0.9, 0.95, 0.99])
# Here, it seems like 95% percentile should be used to drop the outliers
plt.figure(figsize=(12, 6))
plt.title("Distribution of Monetary < 95%")
sns.distplot(RFM[RFM["Monetary"] < 447].Monetary)
RFM = RFM[RFM["Monetary"] < 447]
# # RFM groups
# I have used quantiles for assigning scores for Recency and Monetary.
# * groups are 0-33, 33-66, 66-100 quantiles
# For Frequency I have decided to group them by hand
# * score=1 if the frequency value is 1
# * otherwise, the score will be 2
RFM["R_score"] = pd.qcut(RFM["Recency"], 3, labels=[1, 2, 3]).astype(str)
RFM["M_score"] = pd.qcut(RFM["Monetary"], 3, labels=[1, 2, 3]).astype(str)
RFM["F_score"] = RFM["Frequency"].apply(lambda x: "1" if x == 1 else "2")
RFM["RFM_score"] = RFM["R_score"] + RFM["F_score"] + RFM["M_score"]
# 1. CORE - '123' - most recent, frequent, revenue generating - core customers that should be considered as most valuable clients
# 2. GONE - '311', '312', '313' - gone, one-timers - those clients are probably gone;
# 3. ROOKIE - '111', '112', '113' - just have joined - new clients that have joined recently
# 4. WHALES - '323', '213', '223 - most revenue generating - whales that generate revenue
# 5. LOYAL - '221', '222', '321', '322' - loyal users
# 6. REGULAR - '121', '122', '211', '212', - average users - just regular customers that don't stand out
#
def segment(x):
if x == "123":
return "Core"
elif x in ["311", "312", "313"]:
return "Gone"
elif x in ["111", "112", "113"]:
return "Rookies"
elif x in ["323", "213", "223"]:
return "Whales"
elif x in ["221", "222", "321", "322"]:
return "Loyal"
else:
return "Regular"
RFM["segments"] = RFM["RFM_score"].apply(segment)
RFM["segments"].value_counts(normalize=True) * 100
segmentwise = RFM.groupby("segments").agg(
RecencyMean=("Recency", "mean"),
FrequencyMean=("Frequency", "mean"),
MonetaryMean=("Monetary", "mean"),
GroupSize=("Recency", "size"),
)
segmentwise
font = {"family": "normal", "weight": "normal", "size": 18}
plt.rc("font", **font)
fig = plt.gcf()
ax = fig.add_subplot()
fig.set_size_inches(16, 16)
squarify.plot(
sizes=segmentwise["GroupSize"],
label=segmentwise.index,
color=["gold", "teal", "steelblue", "limegreen", "darkorange", "coral"],
alpha=0.8,
)
plt.title("RFM Segments", fontsize=18, fontweight="bold")
plt.axis("off")
plt.show()
# # Cohort Analysis
#
from operator import attrgetter
joined["order_purchase_timestamp"] = pd.to_datetime(joined["order_purchase_timestamp"])
joined["order_months"] = joined["order_purchase_timestamp"].dt.to_period("M")
joined["cohorts"] = joined.groupby("customer_id")["order_months"].transform("min")
cohorts_data = (
joined.reset_index()
.groupby(["cohorts", "order_months"])
.agg(
ClientsCount=("customer_id", "nunique"),
Revenue=("payment_value", "sum"),
Orders=("order_id", "count"),
)
.reset_index()
)
cohorts_data["periods"] = (cohorts_data.order_months - cohorts_data.cohorts).apply(
attrgetter("n")
) # periods for which the client have stayed
cohorts_data.head()
# Since, majority of our clients are not recurring ones, we can't perform proper cohort analysis on retention and other possible metrics.
# Fortunately, we can analyze dynamics of the bussiness and maybe will be even able to identify some relatively good cohorts that might be used as a prototype (e.g. by marketers).
font = {"family": "normal", "weight": "normal", "size": 12}
plt.rc("font", **font)
fig, (ax1, ax2) = plt.subplots(1, 2, figsize=(24, 6)) # for 2 parrallel plots
cohorts_data.set_index("cohorts").Revenue.plot(ax=ax1)
ax1.set_title("Cohort-wise revenue")
cohorts_data.set_index("cohorts").ClientsCount.plot(ax=ax2, c="b")
ax2.set_title("Cohort-wise clients counts")
# The figure above reveals the dynamics of Revenue and Number of Clients per cohort.
# On the left side we can see Revenue plot and on the right side can see ClientsCount plot.
# Overall, we can come to the next conclusions:
# * dynamics of two graphs are almost identical. Thus, it seems like the Average Order Amount was the same almost for each cohort. It could mean that the only way to get more revenue is to get more clients. Also, we know that we have 97% of non-recurring clients, thus maybe resolving this issue and stimulating customers to comeback would also result in revenue increase
# * I suspect that we don't have the full data for the last several months, because we can see abnormal drop. Thus, these last months shouldn't be taken into considerations
# * Cohort of November-2017 looks like out of trend, since this cohort showed outstanding results. It can be due to Black Friday sales that often happen at Novembers, or maybe during the November of 2017 some experimental marketing campaigns were performed that lead to good results. Thus, this cohort should be investigated by the company in order to identify the reason behind such an outstanding result, and take it into account
ig, (ax1, ax2) = plt.subplots(1, 2, figsize=(24, 6))
(cohorts_data["Revenue"] / cohorts_data["Orders"]).plot(ax=ax1)
ax1.set_title("Average Order Amount per cohort")
sns.boxplot((cohorts_data["Revenue"] / cohorts_data["Orders"]), ax=ax2)
ax2.set_title("Boxplot of the Average Order Amount")
|
import numpy as np
import pandas as pd
import seaborn as sns
import matplotlib.pyplot as plt
from sklearn.preprocessing import LabelEncoder
from sklearn.ensemble import RandomForestRegressor
from sklearn.metrics import r2_score
from sklearn.model_selection import train_test_split, GridSearchCV, cross_val_score
# from bayes_opt import BayesianOptimization
import xgboost as xgb
import warnings
warnings.filterwarnings("ignore")
# ## 1. Data Preprocessing
train = pd.read_csv(
"/kaggle/input/house-prices-advanced-regression-techniques/train.csv"
)
test = pd.read_csv("/kaggle/input/house-prices-advanced-regression-techniques/test.csv")
# combining training set and testing set
df = train.drop(["SalePrice"], axis=1).append(test, ignore_index=True)
# check data types and missing values
df.info()
# 80 predictors in the data frame.
# Data types are string, integer, and float.
# Variables like 'Alley','FireplaceQu','PoolQC','Fence','MiscFeature' contains limited information (too much missing values). But note that some NA values represent "no such equipment" not missing values. (see data description)
# ### 1.1 Missing value analysis
# extract columns contain any null values
missing_values = df.columns[df.isna().any()].tolist()
df[missing_values].info()
# check number of missing values
df[missing_values].isnull().sum()
# Some columns contain limited information (only 198 records in Alley), but note that some of them represent "no such equipment/facility" not missing values (see data description).
# It is true that some predictors there contains a mixing of missing values and "no equipment/facility". "BsmtQual", "BsmtCond" and "BsmtExposure" supposed to be same counts and same number of NAs if no missing values inside.
# To fix this problem I will assume all of the missing values in **some predictors** as "no equipment/facility" if NA is explained in the description file.
# ### 1.2 Missing values imputation (part 1)
df["Alley"] = df["Alley"].fillna("No Access")
df["MasVnrType"] = df["MasVnrType"].fillna("None")
df["BsmtQual"] = df["BsmtQual"].fillna("No Basement")
df["FireplaceQu"] = df["FireplaceQu"].fillna("No Fireplace")
df["GarageType"] = df["GarageType"].fillna("No Garage")
df["PoolQC"] = df["PoolQC"].fillna("No Pool")
df["Fence"] = df["Fence"].fillna("No Fence")
df["MiscFeature"] = df["MiscFeature"].fillna("None")
# the following predictors are linked to the above predictors, should apply a further analysis
# df['BsmtCond'].fillna('No Basement')
# df['BsmtExposure'].fillna('No Basement')
# df['BsmtFinType1'].fillna('No Basement')
# df['BsmtFinType2'].fillna('No Basement')
# df['GarageFinish'].fillna('No Garage')
#
# Now consider other variables (in the comment), there are relationship between variables like 'BsmQual' and 'BsmtCond': only if the value of 'BsmQual' is "no equipment", another should be considered "no equipment" as well.
# check the remaining missing values
missing_values = df.columns[df.isna().any()].tolist()
df[missing_values].isnull().sum()
# clean categorical variables
# fill missing values in 'BsmtCond' column where 'BamtQual' column is 'No Basement'
df.loc[df["BsmtQual"] == "No Basement", "BsmtCond"] = df.loc[
df["BsmtQual"] == "No Basement", "BsmtCond"
].fillna("No Basement")
df.loc[df["BsmtQual"] == "No Basement", "BsmtExposure"] = df.loc[
df["BsmtQual"] == "No Basement", "BsmtExposure"
].fillna("No Basement")
df.loc[df["BsmtQual"] == "No Basement", "BsmtFinType1"] = df.loc[
df["BsmtQual"] == "No Basement", "BsmtFinType1"
].fillna("No Basement")
df.loc[df["BsmtQual"] == "No Basement", "BsmtFinType2"] = df.loc[
df["BsmtQual"] == "No Basement", "BsmtFinType2"
].fillna("No Basement")
df.loc[df["GarageType"] == "No Garage", "GarageFinish"] = df.loc[
df["GarageType"] == "No Garage", "GarageFinish"
].fillna("No Garage")
df.loc[df["GarageType"] == "No Garage", "GarageQual"] = df.loc[
df["GarageType"] == "No Garage", "GarageQual"
].fillna("No Garage")
df.loc[df["GarageType"] == "No Garage", "GarageCond"] = df.loc[
df["GarageType"] == "No Garage", "GarageCond"
].fillna("No Garage")
# clean numerical variables
# fill missing values in 'MasVnrArea' columns where 'MasVnrType' column is 'None'
df.loc[df["MasVnrType"] == "None", "MasVnrArea"] = df.loc[
df["MasVnrType"] == "None", "MasVnrArea"
].fillna(0)
df.loc[df["GarageType"] == "No Garage", "GarageYrBlt"] = df.loc[
df["GarageType"] == "No Garage", "GarageYrBlt"
].fillna(0)
missing_values = df.columns[df.isna().any()].tolist()
df[missing_values].isnull().sum()
# Now most of them only contains one or two missing values.
# Now let's check 'BsmtFinSF1', 'BsmtFinSF2', and 'TotalBsmtSF', since there are only one missing value
df.loc[df["BsmtFinSF1"].isna()].iloc[:, 30:39]
# They are from the same observation index = 2120, since BsmtQual and other variables are labels as "No basement", the Nan values can be labeled as '0'
df.loc[2120] = df.loc[2120].fillna(0)
# check na values in a row
df.loc[2120].isnull().sum()
missing_values = df.columns[df.isna().any()].tolist()
df[missing_values].isnull().sum()
df[missing_values].info()
# ### 1.3 Missing value imputation (part 2)
# Now that most of them only contains 1 or 2 missing values, I will check the distribution of the variables to see what imputation method is more reasonable.
# categorical variables
v1 = [
"MSZoning",
"Utilities",
"Exterior1st",
"Exterior2nd",
"BsmtCond",
"BsmtExposure",
"BsmtFinType2",
"Electrical",
"KitchenQual",
"Functional",
"GarageFinish",
"GarageQual",
"GarageCond",
"SaleType",
]
# quantitative variables
v2 = [x for x in missing_values if x not in v1]
v2.remove("LotFrontage")
for var in v1:
sns.countplot(x=df[var])
plt.show()
for var in v2:
plt.hist(df[var])
plt.title(var)
plt.show()
# Impute categorical variables using **mode**
#
# Impute quantitative variables using **median**
for var in v1:
df[var].fillna(df[var].mode()[0], inplace=True)
for var in v2:
df[var].fillna(df[var].median(), inplace=True)
# Check the missing values one more time, supposed to only have one variable "LotFrontage"
missing_values = df.columns[df.isna().any()].tolist()
df[missing_values].info()
# ### 1.4 Missing value imputation (step 3)
# Missing imputation using Random Forest(RF)
# Encoding
df = df.apply(
lambda x: pd.Series(
LabelEncoder().fit_transform(x[x.notnull()]), index=x[x.notnull()].index
)
)
df.info()
l_known = df[df.LotFrontage.notnull()]
l_unknown = df[df.LotFrontage.isnull()]
l_ytrain = l_known.iloc[:, 3]
l_xtrain = l_known.drop(columns=l_known.columns[3])
l_xtest = l_unknown.drop(columns=l_unknown.columns[3])
rf = RandomForestRegressor()
rf.fit(l_xtrain, l_ytrain)
pred = rf.predict(l_xtest)
df.loc[(df.LotFrontage.isnull()), "LotFrontage"] = pred
df.info()
# ## 2. Modelling
# ### 2.1 train test split
df = df.drop(["Id"], axis=1)
X = df.iloc[0 : train.shape[0], :]
y = train["SalePrice"]
X_test = df.iloc[train.shape[0] :, :]
X_train, X_val, y_train, y_val = train_test_split(
X, y, random_state=123, train_size=0.8
)
# ### 2.2 Hyperparameter tuning
# Benchmark
model = xgb.XGBRegressor()
model.fit(X_train, y_train)
pred = model.predict(X_val)
r2_score(y_val, pred)
params = {
"max_depth": [3, 5, 7],
"gamma": [0.0, 0.3, 0.5, 0.7],
}
model = xgb.XGBRegressor(learning_rate=0.1)
grid_search = GridSearchCV(estimator=model, param_grid=params, cv=5, n_jobs=-1)
grid_search.fit(X_train, y_train)
# Print the best hyperparameters
grid_search.best_params_, grid_search.best_score_
params = {
"reg_alpha": [x / 10 for x in range(1, 11)],
"reg_lambda": [x / 10 for x in range(1, 11)],
}
model = xgb.XGBRegressor(max_depth=3, learning_rate=0.1)
grid_search = GridSearchCV(estimator=model, param_grid=params, cv=5, n_jobs=-1)
grid_search.fit(X_train, y_train)
grid_search.best_params_, grid_search.best_score_
params = {"learning_rate": [x / 10 for x in range(1, 21)]}
model = xgb.XGBRegressor(max_depth=3, reg_alpha=0.8, reg_lambda=0.5)
grid_search = GridSearchCV(estimator=model, param_grid=params, cv=5, n_jobs=-1)
grid_search.fit(X_train, y_train)
grid_search.best_params_, grid_search.best_score_
# ### 2.3 Model fitting & Validating accuracy
model = xgb.XGBRegressor(max_depth=3, learning_rate=0.1, reg_alpha=0.8, reg_lambda=0.5)
model.fit(X_train, y_train)
pred = model.predict(X_val)
r2_score(y_val, pred)
pred = model.predict(X_test)
output = pd.DataFrame({"Id": test["Id"], "SalePrice": pred})
output.head(5)
output.to_csv("submission.csv", index=False)
|
# Note: This notebook was referece for my self-training from https://www.kaggle.com/mathchi/ab-test-for-real-data/ by [Mehmet A.](https://www.kaggle.com/mathchi)
# Since the original dataset is private, I faked one for running it through. Some row of the data was copied data from originally showed. Others was kind of randomly generated.
import os # accessing directory structure
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# # Data Science Analyze and Present A/B Test Results
# AA company recently introduced a new bidding type, “average bidding”, as an alternative to its exisiting bidding
# type, called “maximum bidding”. One of our clients, --------.com, has decided to test this new feature
# and wants to conduct an A/B test to understand if average bidding brings more conversions than maximum
# bidding.
# In this A/B test, --------.com randomly splits its audience into two equally sized groups, e.g. the test
# and the control group. AA company ad campaign with “maximum bidding” is served to “control group” and
# another campaign with “average bidding” is served to the “test group”.
# The A/B test has run for 1 month and --------.com now expects you to analyze and present the results
# of this A/B test.
# ##### You should answer the following questions in your presentation:
# 1. How would you define the hypothesis of this A/B test?
# 2. Can we conclude statistically significant results?
# 3. Which statistical test did you use, and why?
# 4. Based on your answer to Question 2, what would be your recommendation to client?
# ##### Hints:
# 1. Your presentation should last about 15 minutes, and should be presented in English.
# 2. The ultimate success metric for HotelsForYou.com is Number of Purchases. Therefore, you should focus on Purchase metrics for statistical testing.
# 3. Explain the concept of statistical testing for a non-technical audience.
# 4. The customer journey for this campaign is:
# 1. User sees an ad (Impression)
# 2. User clicks on the website link on the ad (Website Click)
# 3. User makes a search on the website (Search)
# 4. User views details of a product (View Content)
# 5. User adds the product to the cart (Add to Cart)
# 6. User purchases the product (Purchase)
# 5. Use visualizations to compare test and control group metrics, such as Website Click Through Rate, Cost per Action, and Conversion Rates in addition to Purchase numbers.
# 6. If you see trends, anomalies or other patterns, discuss these in your presentation.
# 7. You can make assumptions if needed.
# ## 1. Import libraries
import pandas as pd
import numpy as np
import seaborn as sns
import matplotlib.pyplot as plt
from scipy import stats
from scipy.stats import shapiro
from sklearn.neighbors import LocalOutlierFactor
from scipy.stats import levene
from sklearn.impute import KNNImputer
pd.set_option("display.max_columns", None)
pd.set_option("display.max_rows", None)
# ## 2. Functions
def checkReject(p_value, alpha=0.05):
if p_value > alpha:
print(f"p_value is {p_value} > alpha {alpha}, cannot reject null hypoposis")
elif p_value < alpha:
print("p_value is {p_value} < alpha {alpha}, reject null hypoposis")
# ## 3. Data Preparation
control = pd.read_excel(
"/kaggle/input/ab-case-study/AB_Case_Study.xlsx",
sheet_name="Control Group",
index_col=0,
)
test = pd.read_excel(
"/kaggle/input/ab-case-study/AB_Case_Study.xlsx",
sheet_name="Test Group",
index_col=0,
)
control.head()
test.head()
control.info()
# Categorical Columns
categorical = [
col
for col in control.columns
if (control[col].dtype == "object") | (control[col].dtype == "bool")
]
# Numerical Variables
numerical = [col for col in control.columns if col not in categorical]
numerical
kat_control = control[categorical]
num_control = control[numerical]
kat_control.head()
# If we look at the 12th line, we can see the NaN value.
num_control.head(15)
# Fill the control dataframe values NaN value with KNNImputer.
# Fill the control dataframe values NaN value with KNNImputer.
imputer = KNNImputer(n_neighbors=8)
num_control = pd.DataFrame(
imputer.fit_transform(num_control), columns=num_control.columns
)
num_control.iloc[11:12, :]
# Let's combine categorical and numerical variables with the concept.
control = pd.concat([kat_control, num_control], axis=1)
control.head(3)
# data type conversions from float64 to int64.
# Data Type Conversions
control = control.astype(
{
"# of Impressions": "int64",
"Reach": "int64",
"# of Website Clicks": "int64",
"# of Searches": "int64",
"# of View Content": "int64",
"# of Add to Cart": "int64",
"# of Purchase": "int64",
}
)
test = test.astype(
{
"# of Impressions": "int64",
"Reach": "int64",
"# of Website Clicks": "int64",
"# of Searches": "int64",
"# of View Content": "int64",
"# of Add to Cart": "int64",
"# of Purchase": "int64",
}
)
print(control.shape)
print(test.shape)
print(control.info())
print(test.info())
# ## 4. Preparing Data for Hypothesis Testing (A/B Test)
# Let's create a feature with the help of functions and create a feature for visualization and prepare it.
# add columns "Website Click Through Rate" "Number of Action", "Cost per Action"
control["Website Click Through Rate"] = (
control["# of Website Clicks"] / control["# of Impressions"] * 100
)
test["Website Click Through Rate"] = (
test["# of Website Clicks"] / test["# of Impressions"] * 100
)
control["Number of Action"] = (
control["# of Impressions"]
+ control["# of Website Clicks"]
+ control["# of Searches"]
+ control["# of View Content"]
+ control["# of Add to Cart"]
+ control["# of Purchase"]
)
test["Number of Action"] = (
test["# of Impressions"]
+ test["# of Website Clicks"]
+ test["# of Searches"]
+ test["# of View Content"]
+ test["# of Add to Cart"]
+ test["# of Purchase"]
)
control["Cost per Action"] = control["Spend [USD]"] / control["Number of Action"]
test["Cost per Action"] = test["Spend [USD]"] / test["Number of Action"]
control["Conversion Rate"] = (
control["Number of Action"] / control["# of Website Clicks"] * 100
)
test["Conversion Rate"] = test["Number of Action"] / test["# of Website Clicks"] * 100
control_nop = pd.DataFrame(control["# of Purchase"])
test_nop = pd.DataFrame(test["# of Purchase"])
Group_A = control.loc[:, "# of Purchase":"Conversion Rate"].drop(
columns="Number of Action"
)
Group_A["Group"] = "A (MaximumBidding)"
Group_B = test.loc[:, "# of Purchase":"Conversion Rate"].drop(
columns="Number of Action"
)
Group_B["Group"] = "B (AverageBidding)"
AB = pd.concat([Group_A, Group_B])
AB.head(3)
# Let's look at the numbers A and B.
AB["Group"].value_counts()
# ### 4.1. Website Click Through Rate (CTR)
# - It is a rate that shows how often the users who saw the ad CLICK the ad.
# - Number of Ad Clicks / Number of Ad Displays
# - Example: 5 clicks, CTR at 100 impressions = 5%
# - ##### Use visualizations to compare test and control group metrics, such as Website Click Through Rate, Cost per Action, and Conversion Rates in addition to Purchase numbers.
cols = [
"Website Click Through Rate",
"Cost per Action",
"Conversion Rate",
"# of Purchase",
]
fig, axarr = plt.subplots(2, 2, figsize=(18, 9))
for i in [0, 1]:
for j in [0, 1]:
for c in cols:
sns.boxplot(y=c, x="Group", hue="Group", data=AB, ax=axarr[i][j])
cols.remove(c)
break
# Let's create our df for AB test.
df_AB = pd.concat([control_nop, test_nop], axis=1)
df_AB.columns = ["A", "B"]
df_AB.head(3)
# ## 5. Two Independent Sample T Test
# ### 5.1. Assumption Check
# #### 5.1.1. Normality Assumption (**Shapiro Test**)
# * **H0**: Normal distribution assumption is provided.
# * **H1**: ... not provided.
p_value = shapiro(df_AB.A)[1]
checkReject(p_value)
p_value = shapiro(df_AB.B)[1]
checkReject(p_value)
# ### 5.1.2. Variance Homogeneity Assumption (**Levene Test**)
# * **H0** : Variances are homogeneous.
# * **H1** : Variances are not homogeneous.
# **H0** : Varyanslar homojendir.
# **H1** : Varyanslar homojen değildir.
p_value = stats.levene(df_AB.A, df_AB.B)[1]
checkReject(p_value)
# **Comment:** Since the pvalue = 0.36> 0.05, the H0 hypothesis, that is, **the variances were not statistically rejected as homogeneous.**
# ### 5.1.3. Nonparametric Independent Two-Sample T Test (Mann–Whitney U test)
# - Since the normality of the distribution of the data set in which Average Bidding (Test Group) was measured was rejected, the [NonParametric](https://machinelearningmastery.com/nonparametric-statistical-significance-tests-in-python/) Independent Two-Sample T Test was applied for the Hypothesis Test.
# **Hypothesis:**
# * **H0** : When it comes to Maximum Bidding and Average Bidding, there is no statistically significant difference between the purchasing amounts of the two groups. ($\mu_1 = \mu_2$)
# * **H1** : ... there is a difference ($\mu_1 \neq \mu_2$)
p_value = stats.mannwhitneyu(df_AB.A, df_AB.B)[1]
checkReject(p_value)
|
# The main goal of this notebook is provide step by step data analysis, data preprocessing and implement various machine learning tasks. The goal is not just to build a model which gives better results but also to learn various analysis and modeling techniques in the process of building the best model.
# import the required packages
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
import seaborn as sns
import matplotlib.pyplot as plt
sns.set(color_codes=True)
import os
# load train and test data
train_data = pd.read_csv("../input/home-data-for-ml-course/train.csv")
test_data = pd.read_csv("../input/home-data-for-ml-course/test.csv")
train_data.head()
print(
"In training data, there are ",
train_data.shape[0],
" records and ",
train_data.shape[1],
" columns",
)
print(
"In test data, there are ",
test_data.shape[0],
" records and ",
test_data.shape[1],
" columns",
)
# Main data analysis steps:
# remove duplicates,outliers
# remove irrelelvant observations
# Major tasks of data preprocessing
# 1. Handling misssing values
# 2. Handling categorical variables
# 3. Handling outliers
train_data.describe()
# Remove unnnecessary fields
# The column id doesnt provide any additional informaion so we can drop this column
train_data = train_data.drop(["Id"], axis=1)
test_data = test_data.drop(["Id"], axis=1)
y = train_data["SalePrice"]
del train_data["SalePrice"]
# Check if there are any duplicate records
sum(train_data.duplicated())
# # **Handling Missing Data**
train_missing_cols = list(train_data.columns[train_data.isnull().any()])
test_missing_cols = list(test_data.columns[test_data.isnull().any()])
print(
"There are ",
len(train_missing_cols),
" columns with missing values in training data",
)
print("There are ", len(test_missing_cols), " columns with missing values in test data")
train_data.info()
test_data.info()
# percentage of missing values in train data
train_mis_cols = pd.Series(train_data.isnull().sum()) * 100 / train_data.shape[0]
train_mis_cols = train_mis_cols[train_mis_cols > 0]
train_mis_cols
# percentage of missing values in test data
test_mis_cols = pd.Series(test_data.isnull().sum()) * 100 / test_data.shape[0]
test_mis_cols = test_mis_cols[test_mis_cols > 0]
test_mis_cols
# from the above results notice that Alley,FireplaceQu,PoolQC,Fence,MiscFeature have more than 50% missing data. So we can ignore this columns for training the model.
# Theoretically if there are more than 30% of missing values then those columns are said to be ignored
train_data = train_data.drop(
["Alley", "FireplaceQu", "PoolQC", "Fence", "MiscFeature"], axis=1
)
test_data = test_data.drop(
["Alley", "FireplaceQu", "PoolQC", "Fence", "MiscFeature"], axis=1
)
# There will be a relation between lot frontage and lot area. Lets get the square root of lot area and check if it has any correlation with the lot frontage
test_data["SqrtLotArea"] = np.sqrt(test_data["LotArea"])
train_data["SqrtLotArea"] = np.sqrt(train_data["LotArea"])
print(test_data["LotFrontage"].corr(test_data["SqrtLotArea"]))
print(train_data["LotFrontage"].corr(train_data["SqrtLotArea"]))
# From the above results we can see that there is a linear relation between square root of lot area and lot frontage. So lets replace missin vlues of lot frontage with lot area.
test_data.LotFrontage[test_data["LotFrontage"].isnull()] = test_data.SqrtLotArea[
test_data["LotFrontage"].isnull()
]
train_data.LotFrontage[train_data["LotFrontage"].isnull()] = train_data.SqrtLotArea[
train_data["LotFrontage"].isnull()
]
del test_data["SqrtLotArea"]
del train_data["SqrtLotArea"]
def get_counts(data, column):
return data[column].value_counts()
def impute_values(data, column, value):
data.loc[data[column].isnull(), column] = value
print(get_counts(test_data, "MasVnrType"), len(test_data))
print(get_counts(train_data, "MasVnrType"), len(train_data))
# As most the values are none lets replace missing values with None
impute_values(test_data, "MasVnrType", "None")
impute_values(train_data, "MasVnrType", "None")
print(get_counts(test_data, "MasVnrArea"), len(test_data))
print(get_counts(train_data, "MasVnrArea"), len(train_data))
impute_values(test_data, "MasVnrArea", 0.0)
impute_values(train_data, "MasVnrArea", 0.0)
print(get_counts(train_data, "Electrical"), len(train_data))
impute_values(test_data, "Electrical", "SBrkr")
impute_values(train_data, "Electrical", "SBrkr")
def weightedAvg(data, col):
tmp = get_counts(data, col)
return sum(tmp.index * tmp.values) / sum(tmp.values)
impute_values(train_data, "GarageYrBlt", round(weightedAvg(train_data, "GarageYrBlt")))
impute_values(test_data, "GarageYrBlt", round(weightedAvg(test_data, "GarageYrBlt")))
get_counts(train_data, "GarageType")
impute_values(train_data, "GarageType", "Attchd")
impute_values(test_data, "GarageType", "Attchd")
train_data = train_data.replace(
{"ExterQual": {"Ex": 5, "Gd": 4, "TA": 3, "Fa": 2, "Po": 1, np.NaN: 0}}
)
test_data = test_data.replace(
{"ExterQual": {"Ex": 5, "Gd": 4, "TA": 3, "Fa": 2, "Po": 1, np.NaN: 0}}
)
train_data = train_data.replace(
{"ExterCond": {"Ex": 5, "Gd": 4, "TA": 3, "Fa": 2, "Po": 1, np.NaN: 0}}
)
test_data = test_data.replace(
{"ExterCond": {"Ex": 5, "Gd": 4, "TA": 3, "Fa": 2, "Po": 1, np.NaN: 0}}
)
train_data = train_data.replace(
{"BsmtQual": {"Ex": 5, "Gd": 4, "TA": 3, "Fa": 2, "Po": 1, np.NaN: 0}}
)
test_data = test_data.replace(
{"BsmtQual": {"Ex": 5, "Gd": 4, "TA": 3, "Fa": 2, "Po": 1, np.NaN: 0}}
)
train_data = train_data.replace(
{"BsmtCond": {"Ex": 5, "Gd": 4, "TA": 3, "Fa": 2, "Po": 1, np.NaN: 0}}
)
test_data = test_data.replace(
{"BsmtCond": {"Ex": 5, "Gd": 4, "TA": 3, "Fa": 2, "Po": 1, np.NaN: 0}}
)
train_data = train_data.replace(
{"BsmtExposure": {"Gd": 4, "Av": 3, "Mn": 2, "No": 1, np.NaN: 0}}
)
test_data = test_data.replace(
{"BsmtExposure": {"Gd": 4, "Av": 3, "Mn": 2, "No": 1, np.NaN: 0}}
)
train_data = train_data.replace(
{
"BsmtFinType1": {
"GLQ": 6,
"ALQ": 5,
"BLQ": 4,
"Rec": 3,
"LwQ": 2,
"Unf": 1,
np.NaN: 0,
}
}
)
test_data = test_data.replace(
{
"BsmtFinType1": {
"GLQ": 6,
"ALQ": 5,
"BLQ": 4,
"Rec": 3,
"LwQ": 2,
"Unf": 1,
np.NaN: 0,
}
}
)
train_data = train_data.replace(
{
"BsmtFinType2": {
"GLQ": 6,
"ALQ": 5,
"BLQ": 4,
"Rec": 3,
"LwQ": 2,
"Unf": 1,
np.NaN: 0,
}
}
)
test_data = test_data.replace(
{
"BsmtFinType2": {
"GLQ": 6,
"ALQ": 5,
"BLQ": 4,
"Rec": 3,
"LwQ": 2,
"Unf": 1,
np.NaN: 0,
}
}
)
train_data = train_data.replace(
{"HeatingQC": {"Ex": 5, "Gd": 4, "TA": 3, "Fa": 2, "Po": 1, np.NaN: 0}}
)
test_data = test_data.replace(
{"HeatingQC": {"Ex": 5, "Gd": 4, "TA": 3, "Fa": 2, "Po": 1, np.NaN: 0}}
)
train_data = train_data.replace(
{"KitchenQual": {"Ex": 5, "Gd": 4, "TA": 3, "Fa": 2, "Po": 1, np.NaN: 0}}
)
test_data = test_data.replace(
{"KitchenQual": {"Ex": 5, "Gd": 4, "TA": 3, "Fa": 2, "Po": 1, np.NaN: 0}}
)
train_data = train_data.replace(
{
"Functional": {
"Typ": 8,
"Min1": 7,
"Min2": 6,
"Mod": 5,
"Maj1": 4,
"Maj2": 3,
"Sev": 2,
"Sal": 1,
np.NaN: 0,
}
}
)
test_data = test_data.replace(
{
"Functional": {
"Typ": 8,
"Min1": 7,
"Min2": 6,
"Mod": 5,
"Maj1": 4,
"Maj2": 3,
"Sev": 2,
"Sal": 1,
np.NaN: 0,
}
}
)
train_data = train_data.replace(
{"FireplaceQu": {"Ex": 5, "Gd": 4, "TA": 3, "Fa": 2, "Po": 1, np.NaN: 0}}
)
test_data = test_data.replace(
{"FireplaceQu": {"Ex": 5, "Gd": 4, "TA": 3, "Fa": 2, "Po": 1, np.NaN: 0}}
)
train_data = train_data.replace(
{"GarageFinish": {"Fin": 3, "RFn": 2, "Unf": 1, np.NaN: 0}}
)
test_data = test_data.replace(
{"GarageFinish": {"Fin": 3, "RFn": 2, "Unf": 1, np.NaN: 0}}
)
train_data = train_data.replace(
{"GarageQual": {"Ex": 5, "Gd": 4, "TA": 3, "Fa": 2, "Po": 1, np.NaN: 0}}
)
test_data = test_data.replace(
{"GarageQual": {"Ex": 5, "Gd": 4, "TA": 3, "Fa": 2, "Po": 1, np.NaN: 0}}
)
train_data = train_data.replace(
{"GarageCond": {"Ex": 5, "Gd": 4, "TA": 3, "Fa": 2, "Po": 1, np.NaN: 0}}
)
test_data = test_data.replace(
{"GarageCond": {"Ex": 5, "Gd": 4, "TA": 3, "Fa": 2, "Po": 1, np.NaN: 0}}
)
train_data = train_data.replace({"PavedDrive": {"Y": 3, "P": 2, "N": 1, np.NaN: 0}})
test_data = test_data.replace({"PavedDrive": {"Y": 3, "P": 2, "N": 1, np.NaN: 0}})
train_data = train_data.replace(
{"PoolQC": {"Ex": 5, "Gd": 4, "TA": 3, "Fa": 2, "Po": 1, np.NaN: 0}}
)
test_data = test_data.replace(
{"PoolQC": {"Ex": 5, "Gd": 4, "TA": 3, "Fa": 2, "Po": 1, np.NaN: 0}}
)
train_data = train_data.replace(
{"Fence": {"GdPrv": 4, "MnPrv": 3, "GdWo": 2, "MnWw": 1, np.NaN: 0}}
)
test_data = test_data.replace(
{"Fence": {"GdPrv": 4, "MnPrv": 3, "GdWo": 2, "MnWw": 1, np.NaN: 0}}
)
print(pd.crosstab(test_data.MSSubClass, test_data.MSZoning))
print(test_data[test_data["MSZoning"].isnull() == True])
test_data.loc[
(test_data["MSZoning"].isnull()) & (test_data["MSSubClass"] == 20), "MSZoning"
] = "RL"
test_data.loc[
(test_data["MSZoning"].isnull()) & (test_data["MSSubClass"] == 30), "MSZoning"
] = "RM"
test_data.loc[
(test_data["MSZoning"].isnull()) & (test_data["MSSubClass"] == 70), "MSZoning"
] = "RM"
print(get_counts(test_data, "Utilities"), len(test_data))
impute_values(test_data, "Utilities", "AllPub")
print(pd.crosstab(test_data.Exterior1st, test_data.Exterior2nd))
impute_values(test_data, "Exterior1st", "VinylSd")
impute_values(test_data, "Exterior2nd", "VinylSd")
print(pd.crosstab(test_data.BsmtFinSF1, test_data.BsmtFinSF2))
impute_values(test_data, "BsmtFinSF1", 0.0)
impute_values(test_data, "BsmtFinSF2", 0.0)
print(get_counts(test_data, "BsmtUnfSF"), len(test_data))
impute_values(test_data, "BsmtUnfSF", round(weightedAvg(test_data, "BsmtUnfSF")))
test_data = test_data.drop(["TotalBsmtSF"], axis=1)
train_data = train_data.drop(["TotalBsmtSF"], axis=1)
print(get_counts(test_data, "BsmtFullBath"), len(test_data))
impute_values(test_data, "BsmtFullBath", round(weightedAvg(test_data, "BsmtFullBath")))
print(get_counts(test_data, "BsmtHalfBath"), len(test_data))
impute_values(test_data, "BsmtHalfBath", 0.0)
print(get_counts(test_data, "KitchenQual"), len(test_data))
impute_values(test_data, "KitchenQual", "TA")
print(get_counts(test_data, "Functional"), len(test_data))
impute_values(test_data, "Functional", "Typ")
print(get_counts(test_data, "GarageCars"), len(test_data))
impute_values(test_data, "GarageCars", 2.0)
print(get_counts(test_data, "GarageArea"), len(test_data))
impute_values(test_data, "GarageArea", round(weightedAvg(test_data, "GarageArea")))
print(get_counts(test_data, "SaleType"), len(test_data))
impute_values(test_data, "SaleType", "WD")
s = pd.Series(train_data.isnull().sum()) * 100 / train_data.shape[0]
s = s[s > 0]
print(s)
s = pd.Series(test_data.isnull().sum()) * 100 / test_data.shape[0]
s = s[s > 0]
print(s)
# There are no more missing values.
# check correlation between all columns and ignore few columns if they are higly correlated with other columns.
corr_table = train_data.corr(method="pearson")
corr_table[(corr_table > 0.7) | (corr_table < -0.7)]
# from the above results notice that external quality and over quality are highly correlated.
# from the above results notice that year built and garage year built are highly correlated.
# so lets drop garage year built and external quality
del train_data["ExterQual"]
del train_data["GarageYrBlt"]
del test_data["ExterQual"]
del test_data["GarageYrBlt"]
train_data.head()
# # **Handle categorical data**
tmp = train_data.columns.to_series().groupby(train_data.dtypes).groups
print({k.name: v for k, v in tmp.items()})
# feature list for one-hot encoding
oneHotCol = [
"MSSubClass",
"MSZoning",
"Street",
"LotShape",
"LandContour",
"Utilities",
"LotConfig",
"LandSlope",
"Neighborhood",
"Condition1",
"Condition2",
"BldgType",
"HouseStyle",
"RoofStyle",
"RoofMatl",
"Exterior1st",
"Exterior2nd",
"MasVnrType",
"Foundation",
"Heating",
"CentralAir",
"Electrical",
"Functional",
"GarageType",
"SaleType",
"SaleCondition",
]
# process each column in the list
for cols in oneHotCol:
train_data = pd.concat(
(train_data, pd.get_dummies(train_data[cols], prefix=cols)), axis=1
)
del train_data[cols]
test_data = pd.concat(
(test_data, pd.get_dummies(test_data[cols], prefix=cols)), axis=1
)
del test_data[cols]
# # **Normalize data**
# min-max scaling
train_data = (train_data - train_data.min()) / (train_data.max() - train_data.min())
test_data = (test_data - test_data.min()) / (test_data.max() - test_data.min())
print(train_data.shape)
print(test_data.shape)
# Notice that there are different number of columns in train and test data. Lets just consider the columns which are present in both
for col in train_data.columns:
if col not in test_data.columns:
del train_data[col]
for col in test_data.columns:
if col not in train_data.columns:
del test_data[col]
X = train_data
X_test = test_data
print(get_counts(test_data, "Utilities_AllPub"), len(test_data))
impute_values(test_data, "Utilities_AllPub", 1)
# # **Simple Linear Regression Model**
from sklearn import linear_model
lm = linear_model.LinearRegression()
lm.fit(X, y)
results = lm.predict(X_test)
np.mean((np.log(y.values) - np.log(lm.predict(X))) ** 2)
# # **Ridge regression**
from sklearn import linear_model
ridge = linear_model.Ridge(alpha=1.0)
ridge.fit(X, y)
results = ridge.predict(X_test)
np.mean((np.log(y.values) - np.log(ridge.predict(X))) ** 2)
# # Lasso Regression
las = linear_model.Lasso(alpha=1.0)
las.fit(X, y)
results = las.predict(X_test)
np.mean((np.log(y.values) - np.log(las.predict(X))) ** 2)
las_weight = las.coef_
las_weight = pd.DataFrame({"feature": X.columns, "weight": las_weight})
print(las_weight[las_weight["weight"] == 0])
# delete all the above columns
X_new = X.drop(
[
"MSSubClass_40",
"MSSubClass_90",
"MSZoning_RL",
"LotConfig_Inside",
"LandSlope_Mod",
"Neighborhood_Sawyer",
"Condition1_RRNn",
"BldgType_1Fam",
"RoofStyle_Hip",
"RoofStyle_Shed",
"MasVnrType_Stone",
"CentralAir_Y",
"Electrical_SBrkr",
"Functional_7",
"SaleType_Oth",
],
axis=1,
)
# delete all the above columns
X_test_new = X_test.drop(
[
"MSSubClass_40",
"MSSubClass_90",
"MSZoning_RL",
"LotConfig_Inside",
"LandSlope_Mod",
"Neighborhood_Sawyer",
"Condition1_RRNn",
"BldgType_1Fam",
"RoofStyle_Hip",
"RoofStyle_Shed",
"MasVnrType_Stone",
"CentralAir_Y",
"Electrical_SBrkr",
"Functional_7",
"SaleType_Oth",
],
axis=1,
)
lm = linear_model.LinearRegression()
lm.fit(X_new, y)
results = lm.predict(X_test_new)
np.mean((np.log(y.values) - np.log(lm.predict(X_new))) ** 2)
ridge = linear_model.Ridge(alpha=1.0)
ridge.fit(X_new, y)
results = ridge.predict(X_test_new)
np.mean((np.log(y.values) - np.log(ridge.predict(X_new))) ** 2)
las = linear_model.Lasso(alpha=2.0)
las.fit(X_new, y)
results = las.predict(X_test_new)
np.mean((np.log(y.values) - np.log(las.predict(X_new))) ** 2)
# # **Gridsearchcv for selecting parameters for ridge regression**
from sklearn.model_selection import GridSearchCV
parameters = {
"alpha": [0.001, 0.01, 0.1, 1, 10, 15, 20, 30, 50, 100, 200, 300, 500, 1000]
}
model = linear_model.Ridge()
Ridge_reg = GridSearchCV(model, parameters, scoring="neg_mean_squared_error", cv=5)
Ridge_reg.fit(X_new, y)
print(Ridge_reg.best_estimator_)
best_model = Ridge_reg.best_estimator_
best_model.fit(X_new, y)
results = best_model.predict(X_test_new)
np.mean((np.log(y.values) - np.log(best_model.predict(X_new))) ** 2)
sample_data = pd.read_csv("../input/home-data-for-ml-course/sample_submission.csv")
sample_data["SalePrice"] = results
sample_data.to_csv("submission.csv", index=False)
sample_data
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
import matplotlib.pyplot as plt
import seaborn as sns
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input/"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
# Load data
train_dataset_path = "/kaggle/input/jog-description-and-salary-in-indonesia/train.csv"
test_dataset_path = "/kaggle/input/jog-description-and-salary-in-indonesia/test.csv"
train_dataset_raw = pd.read_csv(train_dataset_path, sep="|")
test_dataset_raw = pd.read_csv(test_dataset_path, sep="|")
all_dataset_path = "/kaggle/input/jog-description-and-salary-in-indonesia/all.csv"
all_dataset_raw = pd.read_csv(all_dataset_path, sep="|")
all_dataset_raw.shape
all_dataset_raw.head()
# Return info on dataset
all_dataset_raw.info()
# Display number missing values per column
all_dataset_raw.isna().sum()
target_column = "salary"
train_data = train_dataset_raw.copy()
test_data = test_dataset_raw.copy()
all_data = all_dataset_raw.copy()
all_data
all_data.loc[:, ["salary_currency"]].groupby(by="salary_currency").size()
all_dataset_raw.loc[all_dataset_raw["salary"].isna(), :].index
# Outlier remover
all_data_low = all_data[target_column].quantile(0.01)
all_data_high = all_data[target_column].quantile(0.99)
all_data[target_column] = all_data[target_column][
(all_data_low < all_data[target_column]) & (all_data_high > all_data[target_column])
]
all_data[target_column]
# Null remover
all_data = all_dataset_raw.drop(
index=all_dataset_raw.loc[all_dataset_raw["salary"].isna(), :].index
)
all_data = all_data.reset_index().drop(columns=["id", "index"])
# Check the count for each category in the "gender" columnall_data
# Check the count for each category in the "career_level" column
all_data["career_level"].value_counts()
# Create a countplot to visualize the count of each category in the gender column.
sns.countplot(data=all_data, x="career_level")
plt.title("Number of Workers per level")
plt.ylabel("Number of Workers")
plt.show()
# Check the mean annual mileage for the different driving experience groups
all_data.groupby(by="career_level")["salary"].mean()
|
a = 2
print(a)
type(a)
b = 3.4
print(b)
type(b)
c = "abc"
print(c)
type(c)
# **Variable with number**
# **interger , floating , complex numbar**
d = 3 + 4j
print(d)
type(d)
# **Working with numerical variable**
Gross_profit = 30
Revenue = 100
Gross_profit_margin = (Gross_profit / Revenue) * 100
print(Gross_profit_margin)
type(Gross_profit_margin)
RevA = 8
Revenue = Revenue + RevA
Revenue
# **Variable with String**
address = "sdjhdghdff 1/fvfdjgssff fsdh"
print(address)
type(address)
# **Variable with boolean**
a = 5
b = 10
a < b
# **multipal assignment variable**
a, b, c = "sdgfgh", 23, 23.4
print(a)
print(b)
print(c)
|
# # Setup
import os
import gc
import time
import warnings
gc.enable()
warnings.filterwarnings("ignore")
import numpy as np
import pandas as pd
pd.set_option("display.max_columns", None)
pd.set_option("display.precision", 4)
import matplotlib.pyplot as plt
import seaborn as sns
SEED = 23
os.environ["PYTHONHASHSEED"] = str(SEED)
np.random.seed(SEED)
from sklearn.model_selection import KFold, StratifiedKFold
from sklearn.ensemble import ExtraTreesRegressor, ExtraTreesClassifier
from sklearn.metrics import mean_squared_log_error, roc_auc_score
DATA_DIR = "/kaggle/input/playground-series-s3e11"
train = pd.read_csv(f"{DATA_DIR}/train.csv")
test = pd.read_csv(f"{DATA_DIR}/test.csv")
sample_sub = pd.read_csv(f"{DATA_DIR}/sample_submission.csv")
original = pd.read_csv("/kaggle/input/media-campaign-cost-prediction/train_dataset.csv")
# # Overview
train.info()
train.sample()
original.sample()
original.shape, train.shape, test.shape, sample_sub.shape
original.isna().sum().sum(), train.isna().sum().sum(), test.isna().sum().sum()
train.drop("id", axis=1, inplace=True)
test.drop("id", axis=1, inplace=True)
TARGET = "cost"
features = list(test.columns)
# # Feature and Target distributions
plt.figure(figsize=(10, 4))
plt.suptitle("Train vs Original target distribution", fontsize=14)
sns.kdeplot(x=TARGET, data=train, cut=0, label="train")
sns.kdeplot(x=TARGET, data=original, cut=0, label="original")
plt.legend(fontsize="x-small")
plt.show()
train.nunique()
continuous_features = [
"store_sales(in millions)",
"gross_weight",
"units_per_case",
"store_sqft",
]
discrete_features = [
"unit_sales(in millions)",
"total_children",
"num_children_at_home",
"avg_cars_at home(approx).1",
]
categorical_features = [
"recyclable_package",
"low_fat",
"coffee_bar",
"video_store",
"salad_bar",
"prepared_food",
"florist",
]
fig, ax = plt.subplots(2, 2, figsize=(9, 6))
plt.suptitle(
"Train vs Test vs Original distribution - CONTINUOUS FEATURES", fontsize=14
)
for i, f in enumerate(continuous_features):
row, col = i // 2, i % 2
ax[row][col].set_title(f"{f}")
sns.kdeplot(x=f, data=train, cut=0, ax=ax[row][col], label="train")
sns.kdeplot(x=f, data=test, cut=0, ax=ax[row][col], label="test")
sns.kdeplot(x=f, data=original, cut=0, ax=ax[row][col], label="original")
ax[row][col].legend(fontsize="small")
plt.tight_layout()
plt.show()
print("Train vs Test vs Original distribution - DISCRETE NUMERICAL FEATURES\n")
for f in discrete_features:
counts_df = pd.DataFrame()
counts_df["original"] = np.round(
original[f].value_counts(normalize=True).sort_index(), 4
)
counts_df["train"] = np.round(train[f].value_counts(normalize=True).sort_index(), 4)
counts_df["test"] = np.round(test[f].value_counts(normalize=True).sort_index(), 4)
counts_df.index = sorted(train[f].unique())
print(f"{f} - unique values: {sorted(original[f].unique())}\n")
display(counts_df)
print()
print("Train vs Test vs Original distribution - CATEGORICAL FEATURES\n")
for f in categorical_features:
counts_df = pd.DataFrame()
counts_df["original"] = np.round(
original[f].value_counts(normalize=True).sort_index(), 4
)
counts_df["train"] = np.round(train[f].value_counts(normalize=True).sort_index(), 4)
counts_df["test"] = np.round(test[f].value_counts(normalize=True).sort_index(), 4)
counts_df.index = sorted(train[f].unique())
print(f"{f} - unique values: {sorted(original[f].unique())}\n")
display(counts_df)
print()
# ### Fixing datatypes
train[categorical_features] = train[categorical_features].astype("int8")
test[categorical_features] = test[categorical_features].astype("int8")
original[categorical_features] = original[categorical_features].astype("int8")
train[discrete_features] = train[discrete_features].astype("int8")
test[discrete_features] = test[discrete_features].astype("int8")
original[discrete_features] = original[discrete_features].astype("int8")
train[["store_sqft", "units_per_case"]] = train[
["store_sqft", "units_per_case"]
].astype("int")
test[["store_sqft", "units_per_case"]] = test[["store_sqft", "units_per_case"]].astype(
"int"
)
original[["store_sqft", "units_per_case"]] = original[
["store_sqft", "units_per_case"]
].astype("int")
# # Adverserial Validation
def adversarial_validation(first_dataset, second_dataset, model, features):
scores = []
oof_preds = {}
first_dataset["set"] = 0
second_dataset["set"] = 1
composite = pd.concat([first_dataset, second_dataset], axis=0, ignore_index=True)
X, y = composite[features], composite["set"]
cv = StratifiedKFold(n_splits=5, shuffle=True, random_state=SEED)
for fold, (train_idx, val_idx) in enumerate(cv.split(X, y)):
X_train, y_train = X.loc[train_idx], y.iloc[train_idx]
X_val, y_val = X.loc[val_idx], y.iloc[val_idx]
model.fit(X_train, y_train)
val_preds = model.predict_proba(X_val)[:, 1]
oof_preds.update(dict(zip(val_idx, val_preds)))
auc = roc_auc_score(y_val, val_preds)
scores.append(auc)
print(f"Fold #{fold}: AUC = {auc:.5f}")
_ = gc.collect()
oof_preds = pd.Series(oof_preds).sort_index()
print(f"OOF AUC = {roc_auc_score(y, oof_preds):.5f}\n")
model = ExtraTreesClassifier(
n_estimators=150, max_depth=7, n_jobs=-1, random_state=SEED
)
print("Original vs Train")
adversarial_validation(original, train, model, features)
print("Original vs Test")
adversarial_validation(original, test, model, features)
print("Train vs Test")
adversarial_validation(train, test, model, features)
print("Original + Train vs Test")
composite = pd.concat([original, train], axis=0, ignore_index=True)
adversarial_validation(composite, test, model, features)
gc.collect()
# # Baseline
def eval_metric(y_true, y_pred):
return mean_squared_log_error(y_true, y_pred, squared=False)
def cross_val_predict(model, features, num_folds=5, seed=SEED, extended=False):
scores = []
test_preds = {}
oof_preds = {}
X, y = train[features], train[TARGET]
X_test = test[features]
cv = KFold(n_splits=num_folds, shuffle=True, random_state=seed)
for fold, (train_idx, val_idx) in enumerate(cv.split(X, y)):
X_train, y_train = X.loc[train_idx], y.iloc[train_idx]
X_val, y_val = X.loc[val_idx], y.iloc[val_idx]
if extended: # original data added only to training folds
X_train = pd.concat(
[X_train, original[features]], axis=0, ignore_index=True
)
y_train = pd.concat([y_train, original[TARGET]], axis=0, ignore_index=True)
model.fit(X_train, y_train)
val_preds = model.predict(X_val)
oof_preds.update(dict(zip(val_idx, val_preds)))
test_preds[f"fold{fold}"] = model.predict(X_test)
score = eval_metric(y_val, val_preds)
scores.append(score)
print(f"Fold #{fold}: {score:.5f}", end=" | ")
_ = gc.collect()
test_preds = pd.DataFrame.from_dict(test_preds)
test_preds["mean"] = test_preds.mean(axis=1) # mean of fold-wise predictions
oof_preds = pd.Series(oof_preds).sort_index()
print(f"OOF score: {eval_metric(y, oof_preds):.5f}\n")
return oof_preds, test_preds
model = ExtraTreesRegressor(
n_estimators=200, max_depth=12, n_jobs=-1, random_state=SEED
)
# **Training data only:**
_, test_preds_trn = cross_val_predict(model, features)
# **Training data extended using original dataset:**
_, test_preds_ext = cross_val_predict(model, features, extended=True)
# # Submission files
def create_submission_files(test_preds, model_name, notebook="00"):
for col in test_preds.columns:
sub = sample_sub.copy()
sub[TARGET] = test_preds[col]
sub.to_csv(f"{notebook}_{model_name}_{col}.csv", index=False)
create_submission_files(test_preds_trn, "baseline_trn")
create_submission_files(test_preds_ext, "baseline_ext")
|
# # Electricity DayAhead Prices 2022
# This dataset provides hourly day ahead electricity prices for France and interconnections, sourced from the ENTSO-E Transparency Platform, which is a reputable market data provider for European electricity markets. It is valuable resource for businesses, investors, researchers, and energy consumers interested in analyzing and understanding the dynamics of the electricity market with a high level of granularity.
# The dataset includes historical and forecasted day ahead (DAH) electricity prices for France and interconnections (Germany, Italy, Spain, UK, Belgium). The data is presented at an hourly granularity, covering a specific timeframe, and includes information such as hourly electricity prices in Euros per MWh (Megawatt-hour), date, time.
# The dataset is presented in a structured format, such as CSV or Excel, making it easy to manipulate and analyze using various data analysis tools and techniques. It is ideal for conducting research, building predictive models, or gaining insights into the day ahead electricity prices for France and interconnections at an hourly level.
# **Use Case Examples**
# - Developing hourly price forecasting models for France using machine learning algorithms (Deep Learning, Regressions, Random Forests)
# - Analyzing the impact of cross-border electricity trading on hourly prices (EDA)
# - Studying historical hourly trends and patterns in electricity prices for France (Time Series Analysis, LSTM)
# - Building energy pricing models for business planning and strategy with hourly granularity
# - Conducting research on hourly energy market dynamics and trends
# **Dataset Features**
# - Hourly day ahead electricity prices for France
# - Interconnections prices (Italy, Belgium, Germany, Spain, UK)
# - Hourly granularity with date and time information (datetime format)
# - Structured format (e.g., CSV or Excel) for easy data manipulation
import pandas as pd
import matplotlib.pyplot as plt
# # 1. Load the Data
data = pd.read_csv(
"/kaggle/input/electricity-dayahead-prices-entsoe/electricity_dah_prices.csv",
index_col="date", # set date day as index column
parse_dates=True, # use as datetime format
)
# # 2. Quick EDA
data.columns
data.head(10)
data.info()
countries = ["france", "italy", "belgium", "spain", "uk", "germany"]
for country in countries:
data.groupby("date")[country].mean().plot(legend=country)
plt.show()
data[countries].corr()
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
# # 1 Data
# # 1.1 Introduction
# The data preprocessing part is inspired by [SUPER ONE](https://www.kaggle.com/ailobster)'s notebook [Python Titanic Predictions Top 3%](https://www.kaggle.com/code/ailobster/python-titanic-predictions-top-3/notebook), which provides a comprehensive analysis, visualization, and explanation of feature engineering. It is strongly recommended to read this excellent resource.
# In my notebook, I will simply keep the data preprocessing part as simple as possible and focus more on the model.
# ## 1.2 Initialization
import pandas as pd
import numpy as np
import re
from sklearn.ensemble import RandomForestRegressor
# Load datasets
raw_train = pd.read_csv("/kaggle/input/titanic/train.csv")
raw_test = pd.read_csv("/kaggle/input/titanic/test.csv")
# Merge train and test data
all_data = raw_train.merge(raw_test, how="outer")
# ## 1.3 Feature processing
# Generate Ticket_num
all_data["Ticket_num"] = all_data["Ticket"].map(lambda x: re.sub("\D", "", x))
all_data["Ticket_num"] = pd.to_numeric(all_data["Ticket_num"])
# Fill Embarked na with the mode
all_data["Embarked"] = all_data["Embarked"].fillna("S")
# Random forest regressor to fill na of Age
age_df = all_data[["Age", "Pclass", "Sex", "Parch", "SibSp"]]
age_df = pd.get_dummies(age_df)
known_age = age_df[age_df.Age.notnull()].values
unknown_age = age_df[age_df.Age.isnull()].values
y = known_age[:, 0]
X = known_age[:, 1:]
rfr = RandomForestRegressor(random_state=0, n_estimators=100, n_jobs=-1)
rfr.fit(X, y)
predictedAges = rfr.predict(unknown_age[:, 1::])
all_data.loc[(all_data.Age.isnull()), "Age"] = predictedAges
# Fill Fare
fare = all_data.loc[
(all_data["Embarked"] == "S") & (all_data["Pclass"] == 3), "Fare"
].median()
all_data["Fare"] = all_data["Fare"].fillna(fare)
# ## 1.4 Feature engineering
# Extract the titles from Name
titles = set()
for name in all_data["Name"]:
titles.add(name.split(",")[1].split(".")[0].strip())
Title_Dictionary = {
"Capt": "Officer",
"Col": "Officer",
"Major": "Officer",
"Jonkheer": "Master",
"Don": "Royalty",
"Sir": "Royalty",
"Dr": "Officer",
"Rev": "Officer",
"the Countess": "Royalty",
"Mme": "Mrs",
"Mlle": "Miss",
"Ms": "Mrs",
"Mr": "Mr",
"Mrs": "Mrs",
"Miss": "Miss",
"Master": "Master",
"Lady": "Royalty",
"Dona": "Royalty",
}
# Surname frequency
all_data["Surname"] = all_data["Name"].map(lambda name: name.split(",")[0].strip())
all_data["FamilyGroup"] = all_data["Surname"].map(all_data["Surname"].value_counts())
Female_Child_Group = all_data.loc[
(all_data["FamilyGroup"] >= 2)
& ((all_data["Age"] <= 16) | (all_data["Sex"] == "female"))
]
Female_Child_Group = Female_Child_Group.groupby("Surname")["Survived"].mean()
Dead_List = set(Female_Child_Group[Female_Child_Group.apply(lambda x: x == 0)].index)
Male_Adult_Group = all_data.loc[
(all_data["FamilyGroup"] >= 2)
& (all_data["Age"] > 16)
& (all_data["Sex"] == "male")
]
Male_Adult_List = Male_Adult_Group.groupby("Surname")["Survived"].mean()
Survived_List = set(Male_Adult_List[Male_Adult_List.apply(lambda x: x == 1)].index)
all_data.loc[
(all_data["Survived"].isnull())
& (all_data["Surname"].apply(lambda x: x in Dead_List)),
["Sex", "Age", "Title"],
] = ["male", 28.0, "Mr"]
all_data.loc[
(all_data["Survived"].isnull())
& (all_data["Surname"].apply(lambda x: x in Survived_List)),
["Sex", "Age", "Title"],
] = ["female", 5.0, "Miss"]
# FamilySize
all_data["FamilySize"] = all_data["SibSp"] + all_data["Parch"] + 1
# FamilyLabel
def Fam_label(s):
if (s >= 2) & (s <= 4):
return 2
elif ((s > 4) & (s <= 7)) | (s == 1):
return 1
elif s > 7:
return 0
all_data["FamilyLabel"] = all_data["FamilySize"].apply(Fam_label)
# Deck
all_data["Cabin"] = all_data["Cabin"].fillna("Unknown")
all_data["Deck"] = all_data["Cabin"].str.get(0)
# TicketGroup
Ticket_Count = dict(all_data["Ticket"].value_counts())
all_data["TicketGroup"] = all_data["Ticket"].map(Ticket_Count)
def Ticket_Label(s):
if (s >= 2) & (s <= 4):
return 2
elif ((s > 4) & (s <= 8)) | (s == 1):
return 1
elif s > 8:
return 0
all_data["TicketGroup"] = all_data["TicketGroup"].apply(Ticket_Label)
# ## 1.5 Processed Data
# Select columns
all_data = all_data[
[
"Survived",
"Pclass",
"Sex",
"Age",
"Fare",
"Embarked",
"Title",
"FamilyLabel",
"Deck",
"TicketGroup",
]
]
# Split into train and test sets
all_data = all_data[
[
"Survived",
"Pclass",
"Sex",
"Age",
"Fare",
"Embarked",
"Title",
"FamilyLabel",
"Deck",
"TicketGroup",
]
]
all_data = pd.get_dummies(all_data)
processed_train = all_data[all_data["Survived"].notnull()]
processed_test = all_data[all_data["Survived"].isnull()].drop("Survived", axis=1)
# # 2 Model
# ## 2.1 Introduction
# We are going to use [LibFM](http://libfm.org/) as the our model. [PyFM](https://github.com/coreylynch/pyFM) is a Python implementation of LibFM.
# - Factorization Machines, a general approach that mimics most factorization models through feature engineering, combine the generality of feature engineering with the effectiveness of factorization models in estimating interactions between categorical variables of large domains.
# - The implementation uses stochastic gradient descent with adaptive regularization as a learning method. The adaptive regularization adjusts the regularization automatically while training the model parameters.
# ## 2.2 Initialization
# Install PyFM
import pandas as pd
import numpy as np
from scipy.sparse import csr_matrix
from pyfm import pylibfm
from sklearn.preprocessing import MinMaxScaler
from sklearn.feature_selection import SelectKBest
# Get X, y based on the data
X = processed_train.drop(["Survived"], axis=1)
y = processed_train["Survived"]
# Scale the data
scaler = MinMaxScaler()
X = scaler.fit_transform(X)
# Select important features
selector = SelectKBest(k=20)
X = selector.fit_transform(X, y)
# Compress sparse matrix: necessary step for FM
X_array = csr_matrix(X)
np.random.seed(524)
# Fit the FM model
fm_model = pylibfm.FM(
num_factors=20, # After fine-tuning, initial guesses: [5, 10, 20, 50, 100]
num_iter=200, # After fine-tuning, initial guesses: [10, 50, 100, 150, 200, 250]
verbose=True,
task="classification",
initial_learning_rate=0.001, # After fine-tuning, intial guesses: [0.1, 0.01, 0.001]
learning_rate_schedule="optimal",
validation_size=0.1,
k0=True,
k1=True,
init_stdev=0.01,
seed=524,
)
fm_model.fit(X_array, y)
# It is worth mentioning that the threshold for the last step (x > 0.5) is also fine-tuned (just try different thresholds (0.4 ~ 0.7) and submit each result).
# Submission
X_sub = scaler.transform(processed_test)
X_sub = selector.transform(X_sub)
X_sub_array = csr_matrix(X_sub)
y_sub_soft = fm_model.predict(X_sub_array)
y_sub_hard = [int(1) if x > 0.5 else int(0) for x in y_sub_soft] # Threshold
final_data = raw_test.copy()
final_data["Survived"] = y_sub_hard
submission = final_data[["PassengerId", "Survived"]]
submission_data = pd.DataFrame(submission)
submission_data = submission_data.reset_index(drop=True)
submission_data.to_csv("submission.csv", index=False)
|
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
from sklearn.linear_model import Perceptron
from sklearn.metrics import accuracy_score
from sklearn.tree import DecisionTreeClassifier
from sklearn.svm import SVC
from sklearn.linear_model import LinearRegression
from sklearn.linear_model import LogisticRegression
from keras.models import Sequential
from keras.layers import Dense
from sklearn.neural_network import MLPClassifier
dataset = pd.DataFrame(
{
"Age": [79, 69, 73, 75, 81, 77, 80, 85],
"Weight": [310, 240, 258, 300, 200, 217, 241, 264],
}
)
target = np.array([1, 1, 1, 1, -1, -1, -1, -1])
dataset
PLA = Perceptron()
PLA.fit(dataset, target)
y_pred = PLA.predict(dataset)
accuracy = (y_pred == target).mean()
print("Accuracy:", accuracy)
plt.scatter(dataset["Age"], dataset["Weight"], c=target)
plt.plot([70, 90], [300, 200], "-r")
plt.xlabel("Age")
plt.ylabel("Weight")
plt.show()
Tree = DecisionTreeClassifier(max_depth=1)
Tree.fit(dataset, target)
y_pred = Tree.predict(dataset)
Accuracy = accuracy_score(target, y_pred)
print("Accuracy:", Accuracy)
plt.scatter(dataset["Age"], dataset["Weight"], c=target)
xx, yy = np.meshgrid(range(70, 90), range(200, 300))
Z = Tree.predict(np.c_[xx.ravel(), yy.ravel()])
Z = Z.reshape(xx.shape)
plt.contour(xx, yy, Z, colors="red", alpha=0.5)
plt.xlabel("Age")
plt.ylabel("Weight")
plt.show()
svm = SVC(kernel="linear")
svm.fit(dataset, target)
y_pred = svm.predict(dataset)
Accuracy = accuracy_score(target, y_pred)
print("Accuracy:", Accuracy)
plt.scatter(dataset["Age"], dataset["Weight"], c=target)
xx, yy = np.meshgrid(range(70, 90), range(200, 300))
Z = svm.predict(np.c_[xx.ravel(), yy.ravel()])
Z = Z.reshape(xx.shape)
plt.contour(xx, yy, Z, colors="red", alpha=0.5)
plt.xlabel("Age")
plt.ylabel("Weight")
plt.show()
regression = LinearRegression().fit(dataset, target)
y_pred = np.where(regression.predict(dataset) > 0, 1, -1)
Accuracy = np.mean(y_pred == target)
print("Accuracy:", Accuracy)
plt.scatter(dataset["Age"], dataset["Weight"], c=target)
plt.plot(dataset["Age"], regression.predict(dataset), color="red")
plt.xlabel("Age")
plt.ylabel("Weight")
plt.show()
logisticregression = LogisticRegression()
logisticregression.fit(dataset, target)
y_pred = logisticregression.predict(dataset)
plt.scatter(dataset["Age"], dataset["Weight"], c=target)
xx, yy = np.meshgrid(np.linspace(69, 90, num=100), np.linspace(200, 300, num=100))
Z = logisticregression.predict(np.c_[xx.ravel(), yy.ravel()])
Z = Z.reshape(xx.shape)
plt.contour(xx, yy, Z, colors="red", alpha=0.1)
plt.xlabel("Age")
plt.ylabel("Weight")
plt.show()
model = Sequential()
model.add(Dense(1, input_dim=2, activation="sigmoid"))
model.compile(loss="binary_crossentropy", optimizer="adam", metrics=["accuracy"])
model.fit(dataset, target, epochs=1000, batch_size=8, verbose=0)
loss, accuracy = model.evaluate(dataset, target, verbose=0)
print("Accuracy:", accuracy)
clf = MLPClassifier(
hidden_layer_sizes=(10,),
activation="logistic",
solver="sgd",
max_iter=1000,
random_state=0,
)
clf.fit(dataset, target)
plt.scatter(dataset["Age"], dataset["Weight"], c=target)
xx, yy = np.meshgrid(np.linspace(70, 86, 300), np.linspace(200, 311, 300))
Z = clf.predict(np.c_[xx.ravel(), yy.ravel()])
Z = Z.reshape(xx.shape)
plt.contour(xx, yy, Z, colors="red", alpha=0.5)
plt.xlabel("Age")
plt.ylabel("Weight")
plt.show()
|
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
import seaborn as sns
train_csv = pd.read_csv("/kaggle/input/playground-series-s3e12/train.csv")
test_csv = pd.read_csv("/kaggle/input/playground-series-s3e12/test.csv")
train_csv.head()
train_csv.shape
train_csv.describe()
import seaborn as sns
fig, axes = plt.subplots(2, 3, figsize=(12, 5))
feature_names = ["gravity", "ph", "osmo", "cond", "urea", "calc"]
ax = axes.flatten()
for i, feature in enumerate(feature_names):
axes = ax[i]
sns.kdeplot(data=train_csv, x=feature, hue="target", ax=axes)
plt.tight_layout()
sns.scatterplot(data=train_csv, x="calc", y="gravity", hue="target")
# **There is good amount of overlapping , therefore , chances of overfitting is quite high .**
corr_mat = train_csv.corr()["target"].sort_values(ascending=True)
corr_mat
def heatmap(dataset, label=None):
corr = dataset.corr()
plt.figure(figsize=(10, 6), dpi=300)
mask = np.zeros_like(corr)
# print(mask)
mask[np.triu_indices_from(mask)] = True
sns.heatmap(corr, mask=mask, annot=True, annot_kws={"size": 12}, cmap="viridis")
plt.yticks(fontsize=10)
plt.xticks(fontsize=10)
plt.title(f"{label} Dataset Correlation Matrix\n", fontsize=20, weight="bold")
plt.show()
heatmap(train_csv, label="Train")
# OSMO IS LOOKING TO BE A GREAT FEATURE
# # DATA PROCESSING
training_features = ["cond", "calc"]
target_feature = ["target"]
x_train = train_csv[training_features].values
y_train = train_csv[target_feature].values
x_train.shape
y_train.shape
from sklearn.pipeline import Pipeline
from sklearn.preprocessing import StandardScaler
from sklearn.linear_model import LogisticRegression
num_pipeline = Pipeline([("std_scaler", StandardScaler())])
x_train = num_pipeline.fit_transform(x_train)
x_train.shape
x_train[0]
# # MODELLING
log_reg = LogisticRegression().fit(x_train, y_train.ravel())
proba_predictions = log_reg.predict_proba(x_train)[:, 1]
from sklearn.metrics import roc_auc_score
score = roc_auc_score(y_train.ravel(), proba_predictions)
score
# TESTING THE MODEL
from sklearn.model_selection import StratifiedKFold
from sklearn.base import clone
def test_the_model(model):
results = []
scores = []
skfolds = StratifiedKFold(n_splits=20, random_state=42, shuffle=True)
for train_index, test_index in skfolds.split(x_train, y_train.ravel()):
clone_clf = clone(model)
y = y_train.ravel()
X_train_folds = x_train[train_index]
y_train_folds = y[train_index]
X_test_fold = x_train[test_index]
y_test_fold = y[test_index]
clone_clf.fit(X_train_folds, y_train_folds)
y_pred = clone_clf.predict_proba(X_test_fold)[:, 1]
scores.append(roc_auc_score(y_test_fold, y_pred))
return scores
log_regressor = LogisticRegression()
scores = test_the_model(log_regressor)
val_score = np.mean(np.array(scores))
val_score
import xgboost as xgb
xgb_reg = xgb.XGBClassifier().fit(x_train, y_train.ravel())
proba_predictions = xgb_reg.predict_proba(x_train)[:, 1]
proba_predictions
score = roc_auc_score(y_train.ravel(), proba_predictions)
score
xgb_regressor = xgb.XGBClassifier()
val_scores = test_the_model(xgb_regressor)
val_score = np.mean(val_scores)
val_score
# FINETUNING THE XGB PARAMETERS
regressor = xgb.XGBClassifier(
objective="binary:logistic",
eval_metric="auc",
tree_method="exact",
n_jobs=-1,
max_depth=2,
eta=0.01,
n_estimators=100,
).fit(x_train, y_train.ravel())
y_pred = regressor.predict_proba(x_train)[:, 1]
score = roc_auc_score(y_train.ravel(), y_pred)
print("Train set Score : ", score)
xgb_reg = xgb.XGBClassifier(
objective="binary:logistic",
eval_metric="auc",
tree_method="exact",
n_jobs=-1,
max_depth=2,
eta=0.01,
n_estimators=100,
)
val_score = np.mean(test_the_model(xgb_reg))
val_score
# # SUBMISSION
test_values = test_csv[training_features].values
test_val = num_pipeline.fit_transform(test_values)
test_predictions = regressor.predict_proba(test_val)[:, 1]
test_predictions.shape
submission = pd.DataFrame({"id": test_csv["id"].values, "target": test_predictions})
submission.head()
submission.to_csv("submission.csv", index=False)
test_predictions
|
import pandas as pd
import re
import numpy as np
sla = pd.read_excel(
r"../input/shopee-code-league-20/_DA_Logistics/SLA_matrix.xlsx", engine="openpyxl"
)
orders = pd.read_csv(
r"../input/shopee-code-league-20/_DA_Logistics/delivery_orders_march.csv"
)
sla
# 看起來很奇怪,不過從表中,大概可以猜出是一個對照表,而且index是出發地(from),column是目的地(to),然後表裡面是運送時間
# 他的columns看起來是自動建立的,應該是要第一個row拿來做column才對
# row 6-8看起來像是其他資訊,而且只有第一個column有東西,應該是註解之類的,不過要把它祥細看一下
# 看起來column:'Unnamed: 1'才是我要的index
# row:0才是我要的column
for i in sla.iloc[:, 0]:
print(i)
# 確實row 6-8只是註解而已
# 看起來column:'Unnamed: 1'才是我要的index
sla = sla.set_index("Unnamed: 1")
# row:0才是我要的column
sla.columns = sla.iloc[0]
# 設定表格index和column的name,這樣比較看得懂
sla.index.name = "from"
sla.columns.name = "to"
display(sla)
# 把多的內容拿掉
sla = sla.iloc[1:5, 1:5]
display(sla)
# 把表格中的string轉換成int
sla = sla.applymap(lambda x: int(re.match("^(\d+) working days$", x).group(1)))
display(sla)
display(sla.dtypes)
# 把sla的index和column內容都變小寫
sla.index = sla.index.map(lambda x: x.lower())
sla.columns = sla.columns.map(lambda x: x.lower())
display(sla)
# 因為沒有後面重複的地點,所以只用最後一個字
sla.index = sla.index.map(lambda x: x.split()[-1])
sla.columns = sla.columns.map(lambda x: x.split()[-1])
display(sla)
# 到這邊sla整理完成了
orders.head()
# 先整理時間格式
# 以下是範的應對關係
# 1583137548 (Converted to 2020-03-02 4:25:48 PM Local Time)
print(pd.to_datetime(1583137548, unit="s") + pd.Timedelta(hours=8))
# 1583733540 (Converted to 2020-03-09 1:59:00 PM Local Time)
print(pd.to_datetime(1583733540, unit="s") + pd.Timedelta(hours=8))
# 確定這樣轉換是對的,套用到資料上
# 因為時區都是用同一個時區,所以這邊就直接暴力加8小時,不另外設定時區
orders["pick"] = pd.to_datetime(orders["pick"], unit="s") + pd.Timedelta(hours=8)
orders["1st_deliver_attempt"] = pd.to_datetime(
orders["1st_deliver_attempt"], unit="s"
) + pd.Timedelta(hours=8)
orders["2nd_deliver_attempt"] = pd.to_datetime(
orders["2nd_deliver_attempt"], unit="s"
) + pd.Timedelta(hours=8)
display(orders.head())
# 因為題目只看日期,不看時間,把詳細時間丟掉
orders[["pick", "1st_deliver_attempt", "2nd_deliver_attempt"]] = orders[
["pick", "1st_deliver_attempt", "2nd_deliver_attempt"]
].applymap(lambda x: x.date())
orders[["pick", "1st_deliver_attempt", "2nd_deliver_attempt"]] = orders[
["pick", "1st_deliver_attempt", "2nd_deliver_attempt"]
].astype(
"datetime64"
) # 把datatype變回pd datetime object
display(orders.head())
# 整理一下欄位名稱,這樣比較好打
orders.columns = ["orderid", "pick", "st", "nd", "to", "from"]
display(orders.head())
# 整理地址,變成全小寫
orders["to"] = orders["to"].map(lambda x: x.lower())
orders["from"] = orders["from"].map(lambda x: x.lower())
display(orders.head())
# 這邊可以證明地址的最後一個字都是我們sla裡面有的地點
t = orders["to"].map(lambda x: x.split()[-1])
f = orders["from"].map(lambda x: x.split()[-1])
print(t.isin(sla.columns).all()) # sla的columns是to
print(f.isin(sla.index).all()) # sla的index是from
# 只留下要的地名
orders["to"] = orders["to"].map(lambda x: x.split()[-1])
orders["from"] = orders["from"].map(lambda x: x.split()[-1])
display(orders.head())
# 計算時間距離
# 製作一張表,利用查詢可以把所有日期都映射到int,因為假日的部分不計,所以數字不會增加,
# 這邊看不懂沒關係,直接往下拉,看display出來是甚麼東西就會懂了
# 要先知道製作的區間,所以找到整個dataset的日期最大值及最小值,才有辦法知道起訖日期
max_date = orders[["pick", "st", "nd"]].max().max()
min_date = orders[["pick", "st", "nd"]].min().min()
all_dates = pd.date_range(min_date, max_date) # 包含最後一個日期
# 找出星期日
sunday = all_dates[all_dates.weekday == 6]
# 登記國定假日
holiday = pd.DatetimeIndex(["2020-03-08", "2020-03-25", "2020-03-30", "2020-03-31"])
# 2020-03-08 (Sunday);
# 2020-03-25 (Wednesday);
# 2020-03-30 (Monday);
# 2020-03-31 (Tuesday)
# 找出區間內所有不用上班的日子
day_off = sunday.union(holiday)
# 開始製作表
date_delta = pd.Series(index=all_dates, dtype=int)
# 先從0開始計算
delta_value = 0
for date in all_dates:
if not date in day_off:
# 只有工作天的日子才會+1
delta_value += 1
# 紀錄這天的delta value
date_delta[date] = delta_value
display(date_delta)
# 計算工作日的方法
def calaulate_working_days(start, end):
# 因為nd有可能有nat所以要另外處理
if pd.isnull(end):
return np.nan
return date_delta[end] - date_delta[start]
# 下面這個計算時間的方法,理論上可行,可是跑超久,跑不出來,所以我就放棄了
# holiday = pd.DatetimeIndex(['2020-03-08','2020-03-25','2020-03-30','2020-03-31'])
# def calaulate_working_days(start,end):
# if pd.isnull(end):
# return np.nan
# all_days = pd.date_range(start,end)
# all_days = all_days.difference(all_days[[0]])
# all_days = all_days[all_days.weekday != 6]
# all_days = all_days.difference(holiday)
# return all_days.size
# orders['st_delta'] = orders.apply(lambda df: calaulate_working_days(*df[['pick','st']]),axis=1)
# orders['nd_delta'] = orders.apply(lambda df: calaulate_working_days(*df[['st','nd']]),axis=1)
# 把實際日期抽換成delta_value
# 用replace比用apply花較少時間,所以用replace
orders["pick_value"] = orders["pick"].replace(date_delta)
orders["st_value"] = orders["st"].replace(date_delta)
orders["nd_value"] = orders["nd"].replace(date_delta)
# 利用delta_value相減求得兩個事件之間實際距離幾個工作天
orders["st_delta"] = orders["st_value"] - orders["pick_value"] # 從pick到first_attempt
orders["nd_delta"] = (
orders["nd_value"] - orders["st_value"]
) # 從first_attempt到second_attempt
display(orders.head())
# 因為要用replace減少運算時間,所以sla要改成能被replace使用的樣子
stack_sla = sla.stack()
display(stack_sla)
stack_sla.index = stack_sla.index.map(lambda x: ",".join(x))
display(stack_sla)
# 新增一個欄位,能夠利用stack_sla,做replace
orders["route"] = orders["from"] + "," + orders["to"]
# 用replace取代成送貨時間
orders["transport"] = orders["route"].replace(stack_sla)
display(orders.head())
# 依照定義,開始記錄哪些有late
orders["st_late"] = 0
orders["nd_late"] = 0
orders["is_late"] = 0
# first_attempt_time > sla_matrix ---> st_late
orders.loc[orders["st_delta"] > orders["transport"], "st_late"] = 1
# second_attempt - first_attempt > 3 ---> nd_late
orders.loc[orders["nd_delta"] > 3, "nd_late"] = 1
# first_attempt_time > sla_matrix ---> is_late
orders.loc[orders["st_delta"] > orders["transport"], "is_late"] = 1
# second_attempt - first_attempt > 3 ---> is_late
orders.loc[orders["nd_delta"] > 3, "is_late"] = 1
display(orders.query("(st_late==0)&(nd_late==1)").head(20))
display(orders.query("(st_late==1)&(nd_late==0)").head(20))
display(orders.query("(st_late==1)&(nd_late==1)").head(20))
display(orders.query("(st_late==0)&(nd_late==0)").head(20))
orders[["st_late", "nd_late", "orderid"]].groupby(["st_late", "nd_late"]).agg("count")
answer = orders[["orderid", "is_late"]]
print(answer["is_late"].value_counts())
answer
|
# # Introduction
# Recommender systems are a big part of our lives, recommending products and movies that we want to buy or watch. Recommender systems have been around for decades but have recently come into the spotlight.
# In this notebook, We will discuss three types of recommender system: **(1)Association rule learning** (ARL), **(2)content-based** and **(3)collaborative filtering** approaches. In this notebook, we will explain how to build a recommender system with these three methods.
# The functions of a recommender system are to suggest things to the user based on a variety of criteria. These systems forecast the most probable product that customers would buy, and it is interesting to them. Netflix, Amazon, and other businesses employ recommendation algorithms to assist their clients in locating appropriate items or movies.
# Content:
#
# 1. [**Association Rule Learning - ARL**](#1)
# * 1.1 [Data Preprocessing](#2)
# * 1.1.2 [Business Problem](#3)
# * 1.1.3 [Dataset Story](#4)
# * 1.1.4 [Variables](#5)
# * 1.1.5 [Libraries](#6)
# * 1.1.6 [Load and Check Data](#7)
# * 1.1.7 [Outlier Observations](#8)
# * 1.2 [Preparing ARL Data Structure (Invoice-Product Matrix)](#9)
# * 1.3 [Association Rules](#10)
# * 1.4 [Making Product Suggestions to Users at the Shopping Cart Stage](#11)
#
# 1. [**Content-Based Filtering**](#12)
# * 2.1 [Generating TF-IDF Matrix](#13)
# * 2.1.1 [Libraries](#14)
# * 2.1.2 [CountVectorizer](#15)
# * 2.1.3 [tf-idf](#16)
# * 2.2 [Creating Cosine Similarity Matrix](#17)
# * 2.3 [Making Suggestions Based on Similarities](#18)
# * 2.4 [Functionalize All Code of Content-Based Filtering](#19)
#
# 1. [**Collaborative Filtering**](#20)
# * 3.1 [Item-Based Collaborative Filtering](#21)
# * 3.1.1 [Data Preprocessing](#22)
# * 3.1.2 [Creating the User Movie Df](#23)
# * 3.1.3 [Making Item-Based Movie Suggestions](#24)
# * 3.2 [User-Based Collaborative Filtering](#25)
# * 3.2.1 [Data Preprocessing](#26)
# * 3.2.2 [Determining The Movies Watched By The User To Make A Suggestion](#27)
# * 3.2.3 [Accessing Data and Ids of Other Users Watching the Same Movies](#28)
# * 3.2.4 [Identifying Users with the Most Similar Behaviors to the User to Suggest](#29)
# * 3.2.5 [Calculating the Weighted Average Recommendation Score](#30)
# * 3.3 [Model-Based Collaborative Filtering - Matrix Factorization](#31)
# * 3.3.1 [Data Preprocessing](#32)
# * 3.3.2 [Modelling](#33)
# * 3.3.3 [Model Tuning](#34)
# * 3.3.4 [Final Model and Prediction](#35)
#
# 1. [References](#36)
#
#
# # 1. Association Rule Learning
# Association rule learning is a rule-based machine learning approach for finding significant connections between variables in large databases. It is designed to identify strong rules that have been identified in databases using various measures of interest.
# Our aim is to suggest products to users in the product purchasing process by applying association analysis to the online retail II dataset.
# 1. Data Preprocessing
# 2. Preparing ARL Data Structure (Invoice-Product Matrix))
# 3. Association Rules
# 4. Making Product Suggestions to Users at the Shopping Cart Stage
# ## 1.1 Data Preprocessing
# ### 1.1.2 Business Problem
# To suggest products to customers who have reached the basket stage.
# ### 1.1.3 Dataset Story
# * The data set named Online Retail II is a UK-based online sale.
# * Store's sales between 01/12/2009 - 09/12/2011.
# * The product catalog of this company includes souvenirs. promotion can be considered as products.
# * There is also information that most of its customers are wholesalers..
# ### 1.1.4 Variables
# * **InvoiceNo**: Invoice number. The unique number of each transaction, namely the invoice. Aborted operation if it starts with C.
# * **StockCode**: Product code. Unique number for each product.
# * **Description**: Product name
# * **Quantity**: Number of products. It expresses how many of the products on the invoices have been sold.
# * **InvoiceDate**: Invoice date and time.
# * **UnitPrice**: Product price (in GBP)
# * **CustomerID**: Unique customer number
# * **Country**: The country where the customer lives.
# ### 1.1.5 LIBRARIES
### installlation required
#!pip install mlxtend
# libraries
import pandas as pd
pd.set_option("display.max_columns", None)
# pd.set_option('display.max_rows', None)
pd.set_option("display.width", 500)
# çıktının tek bir satırda olmasını sağlar.
pd.set_option("display.expand_frame_repr", False)
from mlxtend.frequent_patterns import apriori, association_rules
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
#
# ### 1.1.6 Load and Check Data
df = pd.read_csv(
"/kaggle/input/online-retail-ii-data-set-from-ml-repository/Year 2010-2011.csv",
encoding="unicode_escape",
)
df.head()
def check_df(dataframe):
print("################ Shape ####################")
print(dataframe.shape)
print("############### Columns ###################")
print(dataframe.columns)
print("############### Types #####################")
print(dataframe.dtypes)
print("############### Head ######################")
print(dataframe.head())
print("############### Tail ######################")
print(dataframe.tail())
print("############### Describe ###################")
print(dataframe.describe().T)
check_df(df)
#
# ### 1.1.7 Outlier Observations
def outlier_thresholds(dataframe, variable):
quartile1 = dataframe[variable].quantile(0.01)
quartile3 = dataframe[variable].quantile(0.99)
interquantile_range = quartile3 - quartile1
up_limit = quartile3 + 1.5 * interquantile_range
low_limit = quartile1 - 1.5 * interquantile_range
return low_limit, up_limit
def replace_with_thresholds(dataframe, variable):
low_limit, up_limit = outlier_thresholds(dataframe, variable)
dataframe.loc[(dataframe[variable] < low_limit), variable] = low_limit
dataframe.loc[(dataframe[variable] > up_limit), variable] = up_limit
def retail_data_prep(dataframe):
dataframe.dropna(inplace=True)
dataframe = dataframe[~dataframe["Invoice"].str.contains("C", na=False)]
dataframe = dataframe[dataframe["Quantity"] > 0]
dataframe = dataframe[dataframe["Price"] > 0]
replace_with_thresholds(dataframe, "Quantity")
replace_with_thresholds(dataframe, "Price")
return dataframe
df = retail_data_prep(df)
#
# ## 1.2 Preparing ARL Data Structure (Invoice-Product Matrix)
# * In this section, we will create a matrix of invoice and products as in the example below.
# EXAMPLE:
# Description NINE DRAWER OFFICE TIDY SET 2 TEA TOWELS I LOVE LONDON SPACEBOY BABY GIFT SET
# Invoice
# 536370 0 1 0
# 536852 1 0 1
# 536974 0 0 0
# 537065 1 0 0
# 537463 0 0 1
# * We will only work on Germany, let's filter.
df_de = df[df["Country"] == "Germany"]
df_de.head(10)
df_de.groupby(["Invoice", "Description"]).agg({"Quantity": "sum"}).head(10)
# * The ouput indicates **Invoice 536527** has these above items.
# * We want to see the description (products) in the columns.
# * And we want to see whether there are products at the intersections of the matrix or not. We can do this with unstack function, we could have used pivot funct instead of it.
df_de.groupby(["Invoice", "Description"]).agg({"Quantity": "sum"}).unstack().iloc[
0:5, 0:5
]
# * If there is a product in the columns, we expect 1, if not 0.
# * Firstly, we will assign 0 to NaN values.
df_de.groupby(["Invoice", "Description"]).agg({"Quantity": "sum"}).unstack().fillna(
0
).iloc[0:5, 0:5]
# * Seconly, If there is a product in the columns, we will convert it to 1.
# applymap will itinirate all cells in the dataframe. apply would only itinirate in row or columns
df_de.groupby(["Invoice", "Description"]).agg({"Quantity": "sum"}).unstack().fillna(
0
).applymap(lambda x: 1 if x > 0 else 0).iloc[0:5, 0:5]
# * Let's turn all these codes into a single function (named create_invoice_product_df).
def create_invoice_product_df(dataframe, id=False):
if id:
return (
dataframe.groupby(["Invoice", "StockCode"])["Quantity"]
.sum()
.unstack()
.fillna(0)
.applymap(lambda x: 1 if x > 0 else 0)
)
else:
return (
dataframe.groupby(["Invoice", "Description"])["Quantity"]
.sum()
.unstack()
.fillna(0)
.applymap(lambda x: 1 if x > 0 else 0)
)
de_inv_pro_df = create_invoice_product_df(df_de)
de_inv_pro_df.head()
# * It doesn't look good, let's make id = True in **create_invoice_product_df** for better looking matrix
def create_invoice_product_df(dataframe, id=True):
if id:
return (
dataframe.groupby(["Invoice", "StockCode"])["Quantity"]
.sum()
.unstack()
.fillna(0)
.applymap(lambda x: 1 if x > 0 else 0)
)
else:
return (
dataframe.groupby(["Invoice", "Description"])["Quantity"]
.sum()
.unstack()
.fillna(0)
.applymap(lambda x: 1 if x > 0 else 0)
)
de_inv_pro_df = create_invoice_product_df(df_de)
de_inv_pro_df.head()
de_inv_pro_df = create_invoice_product_df(df_de, id=True)
# * Let's define a function for checking StockCode number
def check_id(dataframe, stock_code):
product_name = (
dataframe[dataframe["StockCode"] == stock_code][["Description"]]
.values[0]
.tolist()
)
print(product_name)
# * Let's check first stockcode's name **10002**
check_id(df_de, "10002")
#
# ## 1.3. Association Rules
# * We will subtract the probabilities of all possible products being together.
# minumum support value 0.01, we don't want to get below 0.01
# In real life scenarios, this minimum support value is very low.
frequent_itemsets = apriori(de_inv_pro_df, min_support=0.01, use_colnames=True)
frequent_itemsets.sort_values("support", ascending=False).head(10)
rules = association_rules(frequent_itemsets, metric="support", min_threshold=0.01)
rules.sort_values("support", ascending=False).head()
rules.sort_values("lift", ascending=False).head(10)
# * **antecedent support:** probability of the first product
# * **consequent support:** probability of the second product and others
# * **support:** probability of two products (or more) appearing together
# * **confidence:** when product x is bought, the probability of purchasing product y
# * **lift:** when x is taken, the probability of getting y increases by this much (lift)
# ## 1.4 Making Product Suggestions to Users at the Shopping Cart Stage
# Example Product Id: 22492
product_id = "22492"
check_id(df, product_id)
sorted_rules = rules.sort_values("lift", ascending=False)
recommendation_list = []
for i, product in enumerate(sorted_rules["antecedents"]):
for j in list(product):
if j == product_id:
recommendation_list.append(list(sorted_rules.iloc[i]["consequents"])[0])
recommendation_list[0:3]
check_id(df, "21915")
def arl_recommender(rules_df, product_id, rec_count=1):
sorted_rules = rules_df.sort_values("lift", ascending=False)
recommendation_list = []
for i, product in sorted_rules["antecedents"].items():
for j in list(product):
if j == product_id:
recommendation_list.append(list(sorted_rules.iloc[i]["consequents"]))
recommendation_list = list(
{item for item_list in recommendation_list for item in item_list}
)
return recommendation_list[:rec_count]
check_id(df, "23049")
arl_recommender(rules, "22492", 1)
# If want to see two product suggection
arl_recommender(rules, "22492", 2)
# #### Some Notes
# * For example, if I had 10,000 products, I wouldn't be interested in all of them. In this case it should be done at category level
# * When the person adds a product to the cart, what I will suggest should already be clear.
# * I know what to suggest with product X, but if the person has already bought this product, it is necessary to make a correction accordingly. There must be an intermediate control mechanism. At the database level, the userid should be checked. If the person has not bought that product after checking, it is necessary to recommend that product. This cannot be done. There is a fine line you should consider
# # 2. Content-Based Filtering
# * Represent texts mathematically (vectoring texts)
# * Count Vector (word count)
# * TF-IDF
# In this content-based section, we will go through these steps below.
# 1. Creating the TF-IDF Matrix
# 2. Creating the Cosine Similarity Matrix
# 3. Making Suggestions Based on Similarities
# 4. Preparation of the Working Script
# ### Euclidean Distance
# $ d(p, q) = \sqrt \sum_{i = 1}^\infty x_i$
# Ürün içeriklerinin benzerlikleri üzerinden tavsiyeler geliştirilir.
# ### Cosine Similarity
import pandas as pd
pd.set_option("display.max_columns", None)
pd.set_option("display.width", 500)
pd.set_option("display.expand_frame_repr", False)
from sklearn.feature_extraction.text import TfidfVectorizer
from sklearn.metrics.pairwise import cosine_similarity
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# Euclidean Distance
# It finds the distance between two vectors.
# Cosine Similarity
# A metric focused on the similarity of two vectors.
# ## 2.1 Generating TF-IDF Matrix
# Recommendation System Based on Movie Overviews
# ### 2.1.1 Libraries
import pandas as pd
pd.set_option("display.max_columns", None)
pd.set_option("display.width", 500)
pd.set_option("display.expand_frame_repr", False)
from sklearn.feature_extraction.text import TfidfVectorizer
from sklearn.metrics.pairwise import cosine_similarity
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
df2 = pd.read_csv(
"/kaggle/input/movies-metadata/movies_metadata.csv",
sep=";",
encoding="unicode_escape",
low_memory=False,
)
df2.head()
df2["overview"].head()
#
# #### 2.1.2 CountVectorizer
# In the Count operation, the number of times each word occurs in each document is counted. For instace; let's look at the example below. We have four sentences. We will convert all words to the matrix. If the word is in it, it will count 1 or more, othewise 0.
from sklearn.feature_extraction.text import CountVectorizer
corpus = [
"This is the first document.",
"This document is the second document.",
"And this is the third one.",
"Is this the first document?",
]
vectorizer = CountVectorizer()
X = vectorizer.fit_transform(corpus)
vectorizer.get_feature_names()
X.toarray()
#
# #### 2.1.3 tf-idf
# There are normalized numeric representations.
# * **STEP 1:** TF(t) = (Frequency of occurrence of a t term in the relevant document) / (Total number of terms in the document)(term frequency)
# * **Step 2:** IDF(t) = 1 + log_e(Total number of documents + 1) / (number of documents with t term + 1) (inverse document frequency)
# * **Step 3:** TF-IDF = TF(t) * IDF(t)
# * **Step 4:** L2 normalization to TF-IDF values
from sklearn.feature_extraction.text import TfidfVectorizer
vectorizer = TfidfVectorizer(analyzer="word")
X = vectorizer.fit_transform(corpus)
vectorizer.get_feature_names()
X.toarray()
df2["overview"].head()
tfidf = TfidfVectorizer(stop_words="english")
df2["overview"] = df2["overview"].fillna("")
tfidf_matrix = tfidf.fit_transform(df2["overview"])
tfidf_matrix.shape
#
# ## 2.2 Creating Cosine Similarity Matrix
cosine_sim = cosine_similarity(tfidf_matrix, tfidf_matrix)
cosine_sim.shape
cosine_sim[1]
#
# ## 2.3 Making Suggestions Based on Similarities
indices = pd.Series(df2.index, index=df2["title"])
indices = indices[~indices.index.duplicated(keep="last")]
indices.shape
indices[:10]
# showing the index number of the films
indices["The American President"]
movie_index = indices["The American President"]
# Cosine similarities of The American President movie and other movies.
cosine_sim[movie_index]
similarity_scores = pd.DataFrame(cosine_sim[movie_index], columns=["score"])
movie_indices = similarity_scores.sort_values("score", ascending=False)[1:11].index
df2["title"].iloc[movie_indices]
#
# ## 2.4 Functionalize All Code of Content-Based Filtering
def content_based_recommender(title, cosine_sim, dataframe):
# generates index
indices = pd.Series(dataframe.index, index=dataframe["title"])
indices = indices[~indices.index.duplicated(keep="last")]
# capturing the index of the title
movie_index = indices[title]
# Calculating similarity scores by title
similarity_scores = pd.DataFrame(cosine_sim[movie_index], columns=["score"])
# the top 10 movies except itself(the movie we chose)
movie_indices = similarity_scores.sort_values("score", ascending=False)[1:11].index
return dataframe["title"].iloc[movie_indices]
# Example
content_based_recommender("The Matrix", cosine_sim, df2)
# Example
content_based_recommender("The Godfather", cosine_sim, df2)
## Cosine Similarity Function, if you want to use it again.
# def calculate_cosine_sim(dataframe):
# tfidf = TfidfVectorizer(stop_words='english')
# dataframe['overview'] = dataframe['overview'].fillna('')
# tfidf_matrix = tfidf.fit_transform(dataframe['overview'])
# cosine_sim = cosine_similarity(tfidf_matrix, tfidf_matrix)
# return cosine_sim
# cosine_sim = calculate_cosine_sim(df)
# content_based_recommender('The Dark Knight Rises', cosine_sim, df)
#
# # 3. Collaborative Filtering
# * Item-Based Collaborative Filtering
# * User-Based Collaborative Filtering
# * Model-Based Collaborative Filtering
# ## 3.1 Item-Based Collaborative Filtering
# * Suggestions are made on item similarity.
# * For instance; there are movies that show the same liking structure as The Lord of The Rings movie.
# ### 3.1.2 Data Preprocessing
import pandas as pd
pd.set_option("display.max_columns", 20)
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
movie = pd.read_csv("/kaggle/input/movielens-20m-dataset/movie.csv")
rating = pd.read_csv("/kaggle/input/movielens-20m-dataset/rating.csv")
df3 = movie.merge(rating, how="left", on="movieId")
df3.head()
movie = pd.read_csv("/kaggle/input/movielens-20m-dataset/movie.csv")
rating = pd.read_csv("/kaggle/input/movielens-20m-dataset/rating.csv")
df3 = movie.merge(rating, how="left", on="movieId")
df3.head()
df3.shape
# * We have 20000797 rows, 6 columns, we don't want to use all of them. let's take some sample from this data.
# * You can make the dataset smaller with this below code. It takes sample from orginal data. Frac is in ration of percentage you desire.
# * df = df.sample(frac =.50)
# ### 3.1.2 Creating the User Movie Df
df3.shape
# movie counts
df3["title"].nunique()
# rating count of each movies
df3["title"].value_counts().head()
rating_counts = pd.DataFrame(df3["title"].value_counts())
# we don't want all ratings, therefore we add trashold. It will not bring under 10000
rare_movies = rating_counts[rating_counts["title"] <= 10000].index
# taking out rare movies from dataframe
common_movies = df3[~df3["title"].isin(rare_movies)]
# all ratings
common_movies.shape
# we have 462 movies now
common_movies["title"].nunique()
# let's pivot it
user_movie_df = common_movies.pivot_table(
index=["userId"], columns=["title"], values="rating"
)
user_movie_df.shape
user_movie_df.head(10)
user_movie_df.columns
# columns count and title count are equal
len(user_movie_df.columns)
common_movies["title"].nunique()
#
# ### 3.1.3 Making Item-Based Movie Suggestions
movie_name = "Matrix, The (1999)"
movie_name = user_movie_df[movie_name]
user_movie_df.corrwith(movie_name).sort_values(ascending=False).head(10)
# Another Example
movie_name = "12 Angry Men (1957)"
movie_name = user_movie_df[movie_name]
user_movie_df.corrwith(movie_name).sort_values(ascending=False).head(10)
# random selection of movies
movie_name = pd.Series(user_movie_df.columns).sample(1).values[0]
movie_name = user_movie_df[movie_name]
user_movie_df.corrwith(movie_name).sort_values(ascending=False).head(10)
# Let's put all the codes in a single script
# script of all codes
def create_user_movie_df():
movie = pd.read_csv("/kaggle/input/movielens-20m-dataset/movie.csv")
rating = pd.read_csv("/kaggle/input/movielens-20m-dataset/rating.csv")
df = movie.merge(rating, how="left", on="movieId")
comment_counts = pd.DataFrame(df["title"].value_counts())
rare_movies = comment_counts[comment_counts["title"] <= 1000].index
common_movies = df[~df["title"].isin(rare_movies)]
user_movie_df = common_movies.pivot_table(
index=["userId"], columns=["title"], values="rating"
)
return user_movie_df
# user_movie_df = create_user_movie_df()
def item_based_recommender(movie_name, user_movie_df):
movie_name = user_movie_df[movie_name]
return user_movie_df.corrwith(movie_name).sort_values(ascending=False).head(10)
item_based_recommender("Fight Club (1999)", user_movie_df)
movie_name = pd.Series(user_movie_df.columns).sample(1).values[0]
item_based_recommender(movie_name, user_movie_df)
# this function helps to find the movie names
def check_film(keyword, user_movie_df):
return [col for col in user_movie_df.columns if keyword in col]
check_film("Str", user_movie_df)
item_based_recommender("Forrest Gump (1994)", user_movie_df)
#
# ## 3.2. User-Based Collaborative Filtering
# Suggestions are made based on user similarities.
# * Step 1: Preparing the Data Set
# * Step 2: Determining the Movies Watched by the User to Suggest
# * Step 3: Accessing Data and Ids of Other Users Watching the Same Movies
# * Step 4: Identifying Users with the Most Similar Behaviors to the User to Suggest
# * Step 5: Calculating the Weighted Average Recommendation Score
# ### 3.2.1. Data Preprocessing
# We have already defined user movie matrix above as named **user_movie_df**. We will use this data, instead of making all the data process again.
user_movie_df
# Let's define a random user
random_user = int(pd.Series(user_movie_df.index).sample(1, random_state=35).values)
# the output random user id
random_user
#
# ### 3.2.2. Determining The Movies Watched By The User To Make A Suggestion
# we selected the random_user's movie here
random_user_df = user_movie_df[user_movie_df.index == random_user]
random_user_df
# Taking out all NaN
movies_watched = random_user_df.columns[random_user_df.notna().any()].tolist()
# if you want to see all the movies that watched by random user, execute this
movies_watched
# Let's check if random user watch 2001: A Space Odyssey (1968)
user_movie_df.loc[
user_movie_df.index == random_user, user_movie_df.columns == "Aladdin (1992)"
]
# how many movies he watched
len(movies_watched)
#
# ### 3.2.3. Accessing Data and Ids of Other Users Watching the Same Movies
# James' movies
movies_watched_df = user_movie_df[movies_watched]
movies_watched_df.head()
# Number of people who watched at least one movie in common with James. 137658 people watched at least on movie, common movies count 191
movies_watched_df.shape
# As we can see above, there are lots of people watched at least one movie in common, but we need to put a threshold here. Common movies count 50
# user_movie_count indicates how many movies each user watched
# notnull gives us binary output (1 or 0) if we don't do that, the ratings count, like 3.0 + 4.0 = 7.0
user_movie_count = movies_watched_df.T.notnull().sum()
# moving user_movie_count in the columns
user_movie_count = user_movie_count.reset_index()
user_movie_count.columns = ["userId", "movie_count"]
user_movie_count.head()
# 40 treshold in common movies
user_movie_count[user_movie_count["movie_count"] > 40].sort_values(
"movie_count", ascending=False
)
# how many people watch the same movies with James (he watched 50 movies)
# there is one person watching the same movies
user_movie_count[user_movie_count["movie_count"] == 50].count()
# let's bring users id watching the same movies
users_same_movies = user_movie_count[user_movie_count["movie_count"] > 40]["userId"]
users_same_movies.head()
users_same_movies.count()
#
# ### 3.2.4. Identifying Users with the Most Similar Behaviors to the User to Suggest
# * We will perform 3 steps:
# * 1. We will aggregate data from James and other users.
# * 2. We will create the correlation df.
# * 3. We will find the most similar finders (Top Users)
final_df = pd.concat(
[
movies_watched_df[movies_watched_df.index.isin(users_same_movies)],
random_user_df[movies_watched],
]
)
final_df.head()
final_df.shape
# We set all user in the columns, but it doesn't look good,therefore, we will make them tidy
final_df.T.corr()
# making above matrix tidy
corr_df = final_df.T.corr().unstack().sort_values().drop_duplicates()
corr_df = pd.DataFrame(corr_df, columns=["corr"])
corr_df.index.names = ["user_id_1", "user_id_2"]
corr_df = corr_df.reset_index()
corr_df.head()
# Users with 65 percent or more correlation with James
top_users = corr_df[(corr_df["user_id_1"] == random_user) & (corr_df["corr"] >= 0.65)][
["user_id_2", "corr"]
].reset_index(drop=True)
top_users = top_users.sort_values(by="corr", ascending=False)
top_users.rename(columns={"user_id_2": "userId"}, inplace=True)
top_users
# lets merge our new table with ratings
rating = pd.read_csv("/kaggle/input/movielens-20m-dataset/rating.csv")
top_users_ratings = top_users.merge(
rating[["userId", "movieId", "rating"]], how="inner"
)
# taking out James from the table
top_users_ratings = top_users_ratings[top_users_ratings["userId"] != random_user]
# We have a problem here. There are two levels in data. One is corr, other one is rating. Someone has high corr but rating is 1.0 with James, another person has low corr, but high rating. Which one should we consider?
top_users_ratings.head()
# * We have a problem here. There are two levels in data. One is corr, other one is rating. Someone has high corr but rating is 1.0 with James, another person has low corr, but high rating. Which one should we consider?
# * I need to make a weighting based on rating and correlation.
# ### 3.2.5. Calculating the Weighted Average Recommendation Score
# * We will create a single score by simultaneously considering the impact of the users most similar to James (correlation) and the rating.
# Calculation of weighted_rating
top_users_ratings["weighted_rating"] = (
top_users_ratings["corr"] * top_users_ratings["rating"]
)
top_users_ratings.head()
# * There is one more problem this above table. There are many ratings given to a movie. We will use groupby for this problem.
recommendation_df = top_users_ratings.groupby("movieId").agg(
{"weighted_rating": "mean"}
)
recommendation_df = recommendation_df.reset_index()
recommendation_df.head()
# there are 8071 movies
recommendation_df[["movieId"]].nunique()
# There are 8071 movies, we can't recommend all these movies. we need a trashold. We do not want to recommend **weighted_score** below **3.5**
movies_to_be_recommend = recommendation_df[
recommendation_df["weighted_rating"] > 3.5
].sort_values("weighted_rating", ascending=False)
movies_to_be_recommend
# list of movies to recommend james
movie = pd.read_csv("/kaggle/input/movielens-20m-dataset/movie.csv")
movies_to_be_recommend.merge(movie[["movieId", "title"]])
# def user_based_recommender():
# import pickle
# import pandas as pd
# # user_movie_df = pickle.load(open('user_movie_df.pkl', 'rb'))
# random_user = int(pd.Series(user_movie_df.index).sample(1, random_state=45).values)
# random_user_df = user_movie_df[user_movie_df.index == random_user]
# movies_watched = random_user_df.columns[random_user_df.notna().any()].tolist()
# movies_watched_df = user_movie_df[movies_watched]
# user_movie_count = movies_watched_df.T.notnull().sum()
# user_movie_count = user_movie_count.reset_index()
# user_movie_count.columns = ["userId", "movie_count"]
# users_same_movies = user_movie_count[user_movie_count["movie_count"] > 20]["userId"]
#
# final_df = pd.concat([movies_watched_df[movies_watched_df.index.isin(users_same_movies)],
# random_user_df[movies_watched]])
#
# corr_df = final_df.T.corr().unstack().sort_values().drop_duplicates()
# corr_df = pd.DataFrame(corr_df, columns=["corr"])
# corr_df.index.names = ['user_id_1', 'user_id_2']
# corr_df = corr_df.reset_index()
#
# top_users = corr_df[(corr_df["user_id_1"] == random_user) & (corr_df["corr"] >= 0.65)][
# ["user_id_2", "corr"]].reset_index(drop=True)
#
# top_users = top_users.sort_values(by='corr', ascending=False)
# top_users.rename(columns={"user_id_2": "userId"}, inplace=True)
# rating = pd.read_csv('/kaggle/input/movielens-20m-dataset/rating.csv')
# top_users_ratings = top_users.merge(rating[["userId", "movieId", "rating"]], how='inner')
# top_users_ratings['weighted_rating'] = top_users_ratings['corr'] * top_users_ratings['rating']
# top_users_ratings = top_users_ratings[top_users_ratings["userId"] != random_user]
#
# recommendation_df = top_users_ratings.groupby('movieId').agg({"weighted_rating": "mean"})
# recommendation_df = recommendation_df.reset_index()
#
# movies_to_be_recommend = recommendation_df[recommendation_df["weighted_rating"] > 3.7].sort_values("weighted_rating", ascending=False)
# movie = pd.read_csv('/kaggle/input/movielens-20m-dataset/movie.csv')
# return movies_to_be_recommend.merge(movie[["movieId", "title"]])
# user_based_recommender()
#
# ## 3.3 Matrix Factorization
# * To fill in the blanks, the weights of the latent features that are assumed to exist for users and movles are found over the existing data and predictions are made for non-existent observations with these weights.
# * Matrix factorization assume; There are some latent factors when users like a movie. These hidden factors are also present in movies.
# 1. Decomposes the user-item matrix into 2 less dimensional matrices.
# 2. It assumes that the transition from two matrices to the user-item matrix occurs with latent factors. We will assume the latent variables.
# 3. The weights of the latent factors are found on the filled observations.
# 4. Empty observations are filled with the weights found.
# * The reason why James watches the movie; the genre of the film, the director or actors of the film, the duration of the film, the language in which the film was shot. While you are liking the movie, there are some factors that you are not aware of. These are called latent factors, features in machine learning.
# **add FORMULA OF MATRIX FAC.**
# * It's an optimization, it's actually a gradient descent work.
# Let's make it more clear.
# * It is assumed that the rating matrix is formed by the product of two factor matrices (dot product).
# * Factor matrices? User latent factors, movie latent factors, are actually two separate matrices.
# * Latent factors? Or latent features? Latent factors or variables.
# * Users and movies are considered to have scores for latent features.
# * These weights (scores) are first found on the existing data and then the empty sections are filled according to these weights.
# #### What are these factors in this data?
# Comedy, horror, adventure, action, thriller, youth, having a specific actor, director.
# ADD TABLES | PICTURE
# $ r_{11} = p_{11} * q_{11} + p_{12} * q_{21} $
# * All p and q are found iteratively over the existing values and then used.
# * Initially, random p and q values and the values in the rating matrix are tried to be estimated.
# * In each iteration, erroneous estimations are arranged and the values in the rating matrix are tried to be approached.
# * For example, if 5 is called 3 in one iteration, the next one is called 4, then 5 is called.
# * Thus, p and q values are filled as a result of a certain iteration.
# * Estimation is made for null observations based on the available p and q.
# #### Some Notes
# * Matrix Factorization vs SVD is not the same
# * SVD (Singular Value Decomposition) is a size reduction method.
# * ALS --> Spark ALS for big data. Only difference ALS make some changes on p and q values.
# ### 3.3.1 Data Preprocessing
# pip install surprise
import pandas as pd
from surprise import Reader, SVD, Dataset, accuracy
from surprise.model_selection import GridSearchCV, train_test_split, cross_validate
pd.set_option("display.max_columns", None)
movie = pd.read_csv("/kaggle/input/movielens-20m-dataset/movie.csv")
rating = pd.read_csv("/kaggle/input/movielens-20m-dataset/rating.csv")
df = movie.merge(rating, how="left", on="movieId")
df.head()
# We reduce the dataset to these four movies in terms of both followability and performance..
movie_ids = [130219, 356, 4422, 541]
movies = [
"The Dark Knight (2011)",
"Cries and Whispers (Viskningar och rop) (1972)",
"Forrest Gump (1994)",
"Blade Runner (1982)",
]
sample_df = df[df.movieId.isin(movie_ids)]
sample_df.shape
sample_df.head()
# creating the user movie dataframe
user_movie_df = sample_df.pivot_table(
index=["userId"], columns=["title"], values="rating"
)
user_movie_df.head()
# The surprise library requires between which numbers it will be. We give 1-5 range.
reader = Reader(rating_scale=(1, 5))
# The data we created in accordance with the data structure of the surprise library
data = Dataset.load_from_df(sample_df[["userId", "movieId", "rating"]], reader)
type(data)
#
# ### 3.3.2 Modelling
trainset, testset = train_test_split(data, test_size=0.25)
svd_model = SVD()
svd_model.fit(trainset)
predictions = svd_model.test(testset)
# predictions
accuracy.rmse(predictions)
cross_validate(svd_model, data, measures=["RMSE", "MAE"], cv=5, verbose=True)
user_movie_df.head()
# Let's guess blade runner( 541) for userid 1
svd_model.predict(uid=1.0, iid=541, verbose=True)
# Let's guess Whispers (356) for userid 1. We guessed for the movie she didn't watc
svd_model.predict(uid=1.0, iid=356, verbose=True)
#
# ### 3.3.3 Model Tuning
# There is the problem of how long I will do the process of changing the values. Should I do this replacement 10 times, like 100 times? So this is a hyperparameter for me that needs to be optimized by the user. How many times will I update the weight? With this question, what will my learning speed be? There is a learning rate that represents the speed of these updates.
# here it is our parameters for the model, epoch and learning rate
param_grid = {"n_epochs": [5, 10], "lr_all": [0.002, 0.005]}
gs = GridSearchCV(
SVD, param_grid, measures=["rmse", "mae"], cv=3, n_jobs=-1, joblib_verbose=True
)
gs.fit(data)
gs.best_score["rmse"]
gs.best_params["rmse"]
#
# ### 3.3.4 Final Model and Prediction
svd_model = SVD(**gs.best_params["rmse"])
data = data.build_full_trainset()
svd_model.fit(data)
user_movie_df.head()
# Let's guess blade runner(541) for userid 1
svd_model.predict(uid=1.0, iid=541, verbose=True)
# Another example Cries and Whispers (356) for user id1.
svd_model.predict(uid=1.0, iid=356, verbose=True)
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
df = pd.read_csv("/kaggle/input/customer-shopping-dataset/customer_shopping_data.csv")
# # **You can find the whole Jupyter Notebook along with a web application for this notebook in the GitHub link [here](https://github.com/Ishrak30/Customer-Spending-Prediction-using-ML-Techniqiues)**
# # **Understanding Data**
df.head(10)
df.describe()
df[["day", "month", "year"]] = df["invoice_date"].str.split("/", expand=True)
print(df.shape)
print(df.columns)
print(df.dtypes)
df.isnull().sum()
categorical_feature = ["gender", "category", "payment_method", "shopping_mall"]
numerical_feature = ["age", "quantity", "month", "year"]
dropping = ["customer_id", "invoice_no", "day", "invoice_date"]
result = ["price"]
df.drop(dropping, axis=1, inplace=True)
df.columns
dt = df[categorical_feature]
for col in dt:
print("Features: ", col, "--", dt[col].unique())
# # **Exploratory Data Analysis**
import seaborn as sns
import plotly.express as px
import matplotlib.pyplot as plt
from matplotlib import style
# ## **Categorical Feature Plot**
# fig, ax = plt.subplots(figsize=(15, 5))
fig = px.histogram(
df,
x="category",
y="price",
color="gender",
labels={"category": "Category", "gender": "Gender"},
)
fig.show()
fig = px.histogram(
df,
x="year",
y="quantity",
color="gender",
labels={"year": "Years", "gender": "Gender", "sum of quantity": "Quantity"},
)
fig.show()
fig = px.histogram(
df,
x="gender",
y="price",
color="gender",
labels={"category": "Category", "gender": "Gender"},
)
fig.show()
# df['shopping_mall'].unique()
fig = px.histogram(
df,
x="shopping_mall",
y="price",
color="gender",
labels={"shopping_mall": "Mall Name", "gender": "Gender"},
)
fig.show()
df_mall_cat = (
df.groupby(["shopping_mall", "category"])["price"]
.sum()
.unstack("category")
.plot(kind="bar", figsize=(15, 8))
)
plt.xlabel("Price")
plt.title("Price for each item in different mall")
plt.ylabel("Mall")
plt.xticks(rotation=45)
plt.show()
# ## Numerical Feature Plot
# how many quantity is bought each year by gender
fig = px.histogram(
df,
x="year",
y="quantity",
color="gender",
labels={"year": "Year", "gender": "Gender"},
)
fig.show()
# payment method by gender every category
fig = px.box(
df,
x="category",
y="payment_method",
color="gender",
labels={
"category": "Categories",
"gender": "Gender",
"payment_method": "Payment Methods",
},
)
fig.show()
# # **Feature Encoding**
from sklearn.preprocessing import LabelEncoder
le = LabelEncoder()
for col in categorical_feature:
df[col] = le.fit_transform(df[col])
df.head()
# # **Correlation Matrix**
##Correlation Matrix using Seaborn
# corr = df.corr()
# #mask for upper triangle
# mask = np.triu(np.ones_like(corr, dtype=bool))
# #matplotlib figure
# fig, ax = plt.subplots(figsize=(11,9))
# #colormap
# cmap = sns.color_palette("coolwarm", as_cmap=True)
# #cmap = sns.diverging_palette(230, 20, as_cmap=True)
# #plot
# dataplot = sns.heatmap(corr, mask=mask, cmap=cmap, vmax=.3, center=0,
# square=True, linewidths=.5, cbar_kws={"shrink": .5})
##Correlation Matrix using Plotly
import plotly.graph_objects as go
import plotly.figure_factory as ff
df_corr = df.corr() # Generate correlation matrix
fig = go.Figure()
fig.add_trace(
go.Heatmap(
x=df_corr.columns,
y=df_corr.index,
z=np.array(df_corr),
colorscale="teal",
text=np.array(df_corr),
texttemplate="%{text}",
)
)
fig.show()
# # Models
# split data
from sklearn.model_selection import train_test_split
# data modelling
from sklearn.linear_model import LinearRegression
from sklearn.metrics import (
confusion_matrix,
accuracy_score,
roc_curve,
classification_report,
)
from sklearn import metrics
y = df["price"]
X = df.drop("price", axis=1)
X_train, X_test, y_train, y_test = train_test_split(
X, y, test_size=0.20, random_state=0
)
# from sklearn.preprocessing import StandardScaler
# sc_std = StandardScaler()
# X_train = sc_std.fit_transform(X_train)
# X_test = sc_std.transform(X_test)
X_train
# ## Working on Models
# ### Linear Regression
m1 = "Linear Regression"
lr = LinearRegression()
model = lr.fit(X_train, y_train)
plt.scatter(lr.predict(X_train), y_train)
plt.xlabel("Predicted value of Y")
plt.ylabel("Real value of Y")
plt.show()
from sklearn.metrics import mean_squared_error
pred = lr.predict(X_test)
rms = mean_squared_error(y_test, pred, squared=False)
print(rms)
print("Training R2 Score - ", lr.score(X_train, y_train) * 100)
print("Test R2 Score - ", lr.score(X_test, y_test) * 100)
# ### Random Forest
from sklearn.ensemble import RandomForestRegressor
m2 = "Random Forest"
rf = RandomForestRegressor(n_estimators=250, n_jobs=-1)
rf.fit(X_train, y_train)
plt.scatter(rf.predict(X_train), y_train)
plt.xlabel("Predicted value of Y")
plt.ylabel("Real value of Y")
plt.show()
print("Training R2 Score - ", rf.score(X_train, y_train) * 100)
print("Test R2 Score - ", rf.score(X_test, y_test) * 100)
# ### Decision Tree
from sklearn.tree import DecisionTreeRegressor
m3 = "Decision Tree"
dtm = DecisionTreeRegressor(max_depth=5, min_samples_split=6, max_leaf_nodes=10)
dtm.fit(X_train, y_train)
plt.scatter(dtm.predict(X_train), y_train)
plt.xlabel("Predicted value of Y")
plt.ylabel("Real value of Y")
plt.show()
print("R2 on train dataset = ", dtm.score(X_train, y_train) * 100)
print("R2 on test dataset = ", dtm.score(X_test, y_test) * 100)
# ### Lasso Regression
from sklearn.linear_model import Lasso, LassoCV
ls_cv = LassoCV(alphas=None, cv=10, max_iter=100000)
ls_cv.fit(X_train, y_train)
alpha = ls_cv.alpha_
alpha
ls = Lasso(alpha=ls_cv.alpha_)
ls.fit(X_train, y_train)
plt.scatter(ls.predict(X_train), y_train)
plt.xlabel("Predicted value of Y")
plt.ylabel("Real value of Y")
plt.show()
print("Train score: ", ls.score(X_train, y_train) * 100)
print("Test score: ", ls.score(X_test, y_test) * 100)
# ### Ridge Regression
from sklearn.linear_model import Ridge, RidgeCV
alphas = np.random.uniform(0, 10, 50)
r_cv = RidgeCV(alphas=alphas, cv=10)
r_cv.fit(X_train, y_train)
alpha = r_cv.alpha_
alpha
ri = Ridge(alpha=r_cv.alpha_)
ri.fit(X_train, y_train)
plt.scatter(ri.predict(X_train), y_train)
plt.xlabel("Predicted value of Y")
plt.ylabel("Real value of Y")
plt.show()
print("Train score: ", ri.score(X_train, y_train) * 100)
print("Test score: ", ri.score(X_test, y_test) * 100)
# # Here I will be using Decision tree for output since it has the best score and does not seem to be overfitting or underfitting.
# Export pickle file for webapp
# import pickle
# filename = 'webapp-pickle-dtm'
# pickle.dump(dtm,open(filename,'wb'))
# model = pickle.load(open(filename, 'rb'))
# user_input=[[0,53,0,4,0,4,10,2021]]
# user_input=np.array(user_input)
# prediction = model.predict(user_input)
# prediction
|
End of preview. Expand
in Data Studio
README.md exists but content is empty.
- Downloads last month
- 11