The dataset viewer is not available for this subset.
Exception: SplitsNotFoundError
Message: The split names could not be parsed from the dataset config.
Traceback: Traceback (most recent call last):
File "/src/services/worker/.venv/lib/python3.9/site-packages/datasets/inspect.py", line 299, in get_dataset_config_info
for split_generator in builder._split_generators(
File "/src/services/worker/.venv/lib/python3.9/site-packages/datasets/packaged_modules/parquet/parquet.py", line 60, in _split_generators
self.info.features = datasets.Features.from_arrow_schema(pq.read_schema(f))
File "/src/services/worker/.venv/lib/python3.9/site-packages/pyarrow/parquet/core.py", line 2325, in read_schema
file = ParquetFile(
File "/src/services/worker/.venv/lib/python3.9/site-packages/pyarrow/parquet/core.py", line 318, in __init__
self.reader.open(
File "pyarrow/_parquet.pyx", line 1470, in pyarrow._parquet.ParquetReader.open
File "pyarrow/error.pxi", line 91, in pyarrow.lib.check_status
OSError: Couldn't deserialize thrift: TProtocolException: Exceeded size limit
The above exception was the direct cause of the following exception:
Traceback (most recent call last):
File "/src/services/worker/src/worker/job_runners/config/split_names.py", line 65, in compute_split_names_from_streaming_response
for split in get_dataset_split_names(
File "/src/services/worker/.venv/lib/python3.9/site-packages/datasets/inspect.py", line 353, in get_dataset_split_names
info = get_dataset_config_info(
File "/src/services/worker/.venv/lib/python3.9/site-packages/datasets/inspect.py", line 304, in get_dataset_config_info
raise SplitsNotFoundError("The split names could not be parsed from the dataset config.") from err
datasets.inspect.SplitsNotFoundError: The split names could not be parsed from the dataset config.Need help to make the dataset viewer work? Make sure to review how to configure the dataset viewer, and open a discussion for direct support.
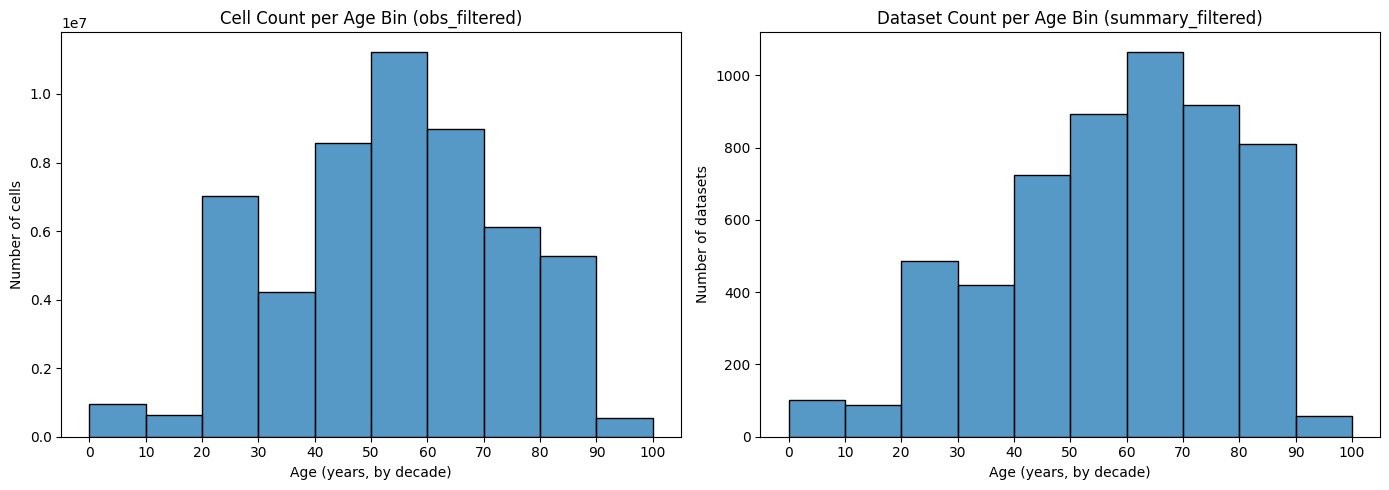
Cellxgene Human Aging Meta Summary
A curated summary of human single-cell RNA-seq datasets from cellxgene, focused on aging and development, with standardized metadata across experiments and assays.
Intended Use
This summary is intended as a scaffold for integrating other curated or custom single-cell datasets (e.g., aging or disease-focused studies) with Cellxgene metadata. Enables exploration, QC, and pre-integration filtering at the experiment level. The seafront package contains code to pull the gene matrices associated with this metadata.
Dataset Description
This dataset provides a summarized table where:
- Each row corresponds to a unique
dataset_id+ simplified assay combination (experiment). - Numeric columns (e.g.
n_measured_vars,raw_sum) are aggregated as medians per dataset. - Categorical metadata (e.g.
cell_type,assay,tissue,disease) are reported as comma-separated unique values. - Ontology term IDs are also preserved and listed uniquely.
- Ambiguous developmental stages are filtered out; valid stages are converted to integer ages (
age_int). - Low-quality experiments (e.g. with median counts < 3000) are excluded.
Data Sources
- Data pulled from the
cellxgene_censusAPI (version 2025-01) usingcellxgene_census.open_soma(). - All human single-cell datasets with interpretable metadata and sufficient quality metrics were included.
Processing Steps
- Assay Simplification: Assay names are normalized into categories like
10x_3_prime,10x_5_prime,BD_WTA,ScaleBio, etc. - Experiment Definition: An
experimentis defined as a unique combination ofdataset_idandassay_simple. - Age Filtering: Text-based developmental stages are mapped to integer age values using a rule-based filter.
- Quality Filtering: Datasets with median raw counts < 3000 are excluded.
- Metadata Summary: Output table generated using a per-experiment groupby summarization of all relevant columns.
Files
cellxgene_human_dataset_overview.parquet: Cleaned and summarized metadata per experiment.- All variable names used in the datasets are saved for future integration compatibility.
Example Columns
experiment,dataset_id,donor_idunique_cell_types_presentunique_tissues_presentunique_assays_present,assay_simplemedian_raw_sum,median_n_measured_varsage_int(if available)
Citation
Please cite the original Cellxgene datasets as per their respective DOIs. This metadata summary is derived and standardized but does not modify raw gene expression data.
- Downloads last month
- 23