Generative Enzyme Design Guided by Functionally Important Sites and Small-Molecule Substrates
Model Architecture
This repository contains code, data and model weights for ICML 2024 paper Generative Enzyme Design Guided by Functionally Important Sites and Small-Molecule Substrates
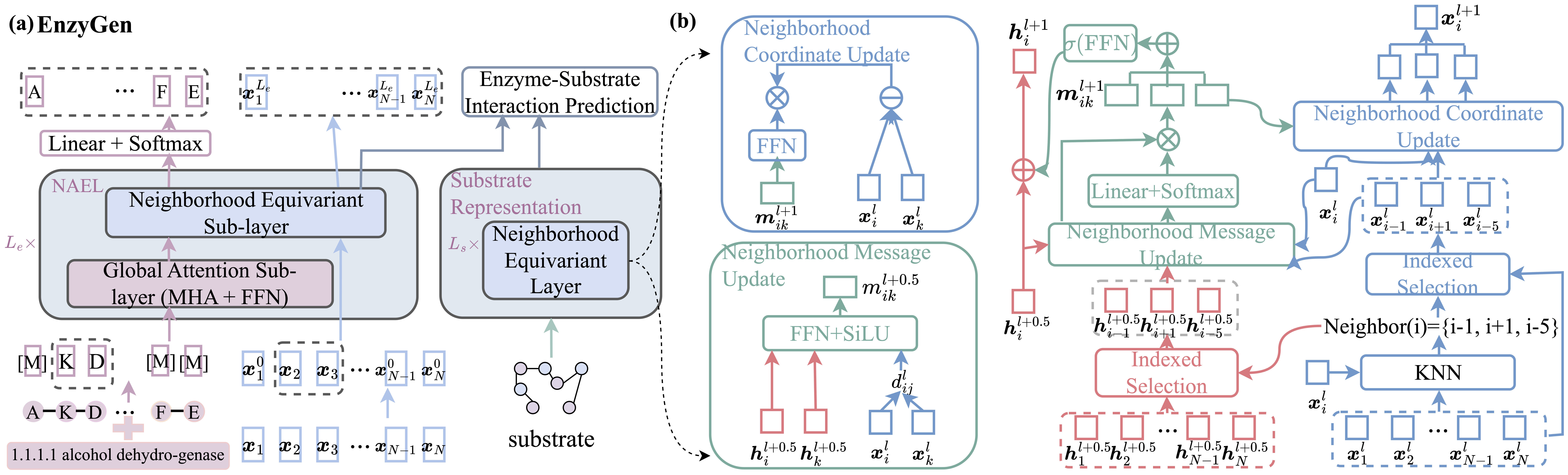
The overall model architecture is shown below:
Usage
1. Clone the Repository
Make sure you have git lfs installed
git clone https://huggingface.co/charlesnovak/EnzyGen
cd EnzyGen
2. Set up the Conda Environment
Make sure you have Conda installed. Then run,
bash setup_conda.sh
conda activate enzygen
3. Prepare Input Data
Modify the provided data/input_example.json
4. Edit infer.sh for Your Task
Make sure paths are correctly provided and the EC numbers for the proteins in the input data are provided
5. Run Inference
bash infer.sh
Outputs
There are 5 items in the outputs directory
- protein.txt refers to the designed protein sequence
- src.seq.txt refers to the ground truth sequences
- pdb.txt refers to the target PDB ID and the corresponding chain
- pred_pdbs refers to the directory of designed pdbs
- tgt_pdbs refers to the directory of target pdbs
Citation
@inproceedings{songgenerative,
title={Generative Enzyme Design Guided by Functionally Important Sites and Small-Molecule Substrates},
author={Song, Zhenqiao and Zhao, Yunlong and Shi, Wenxian and Jin, Wengong and Yang, Yang and Li, Lei},
booktitle={Forty-first International Conference on Machine Learning}
}
Inference Providers
NEW
This model isn't deployed by any Inference Provider.
🙋
Ask for provider support