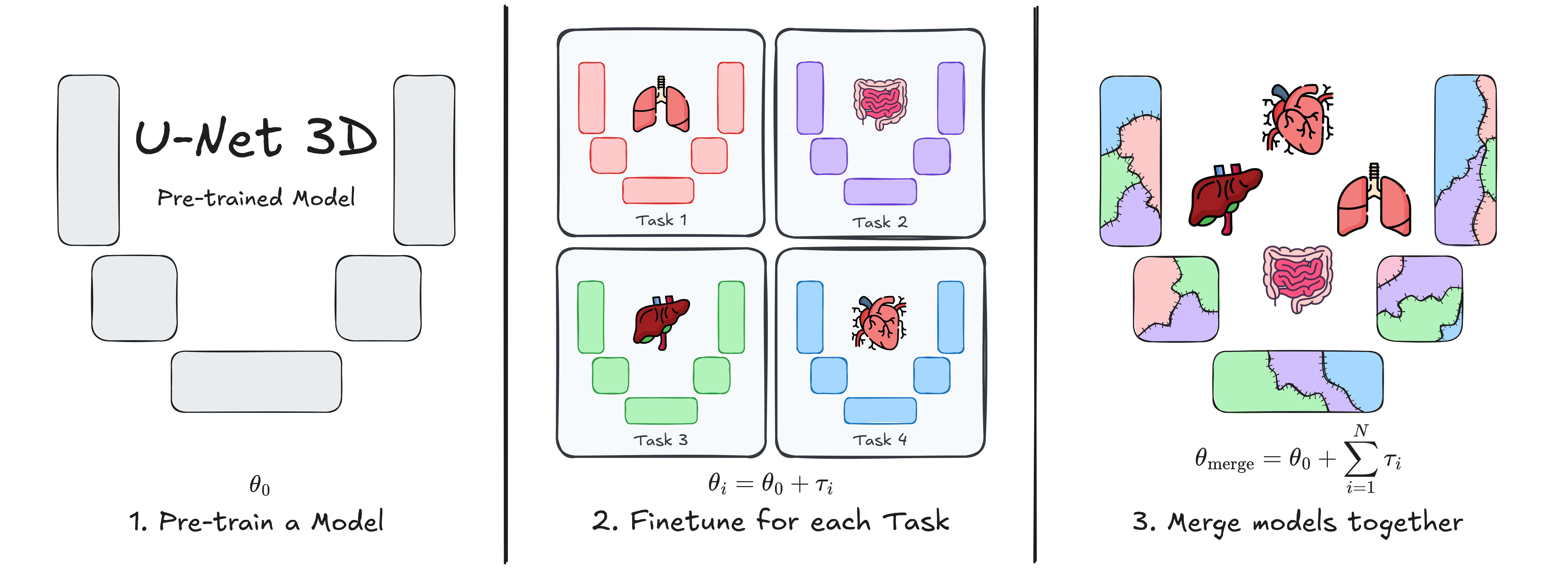
U-Net Transplant: Model Merging for 3D Medical Segmentation
This repository contains the implementation of U-Net Transplant, a framework for efficient model merging in 3D medical image segmentation. Model merging enables the combination of specialized segmentation models without requiring full retraining, offering a flexible and privacy-conscious solution for updating AI models in clinical applications.
Our approach leverages task vectors and encourages wide minima during pre-training to enhance the effectiveness of model merging. We evaluate this method using the ToothFairy2 and BTCV Abdomen datasets with a standard 3D U-Net architecture, demonstrating its ability to integrate multiple specialized segmentation tasks into a single model.
Pretrain and Task Vector Checkpoints
The related checkpoints and task vectors used in the paper will be available from the 23rd June 2025.
How to Run
1. Clone the Repository
git clone [email protected]:LucaLumetti/UNetTransplant.git
cd UNetTransplant
2. Setup Environment
python -m venv env
source env/bin/activate
pip install -r requirements.txt
3. Downloads
Ensure the datasets are downloaded and organized following the nnUNet dataset format.
- BTCV Abdomen: Download Here
- ToothFairy2: Download Here
- AMOS: Download Here
- ZhimingCui: Available upon request from the authors (Paper)
You can also download pretrained checkpoints and task vectors:
#!/bin/bash
BASE_ABDOMEN="https://huggingface.co/Lumett/UNetTransplant/resolve/main/Abdomen"
BASE_TOOTHFAIRY="https://huggingface.co/Lumett/UNetTransplant/resolve/main/ToothFairy"
abdomen_files=(
Pretrain_AMOS.pth
TaskVector_Kidney_Abdomen.pth
TaskVector_Liver_Abdomen.pth
TaskVector_Spleen_Abdomen.pth
TaskVector_Stomach_Abdomen.pth
)
toothfairy_files=(
Pretrain_Cui.pth
TaskVector_Canals_ToothFairy2.pth
TaskVector_Mandible_ToothFairy2.pth
TaskVector_Teeth_ToothFairy2.pth
TaskVector_Pharynx_ToothFairy2.pth
)
echo "🩻 Downloading Abdomen files..."
for file in "${abdomen_files[@]}"; do
wget -c "${BASE_ABDOMEN}/${file}"
done
echo "🦷 Downloading ToothFairy files..."
for file in "${toothfairy_files[@]}"; do
wget -c "${BASE_TOOTHFAIRY}/${file}"
done
4. Running the U-Net Transplant Framework
The main script for running experiments is main.py. It requires specifying the type of experiment and a configuration file that defines dataset, model, optimizer, and training parameters.
Command Structure
python main.py --experiment <EXPERIMENT_TYPE> --config <CONFIG_PATH> [--expname <NAME>] [--override <PARAMS>]
Arguments
--experiment: Specifies the type of experiment to run."PretrainExperiment"→ Pretrains the model from scratch."TaskVectorTrainExperiment"→ Trains a task vector using a pretrained checkpoint.
--config: Path to the configuration file, which defines dataset, model, and training settings.--expname(optional): Custom experiment name. If not provided, the config filename is used.--override(optional): Allows overriding config values at runtime. Example:python main.py --experiment PretrainExperiment --config configs/default.yaml --override DataConfig.BATCH_SIZE=4 OptimizerConfig.LR=0.01
Configuration File
The configuration file defines:
- Dataset (
DataConfig): Path, batch size, patch size, and datasets used. - Model (
BackboneConfig&HeadsConfig): Architecture, checkpoints, and initialization. - Optimizer (
OptimizerConfig): Learning rates, weight decay, and momentum. - Loss Function (
LossConfig): Defines the loss function used. - Training (
TrainConfig): Number of epochs, checkpoint saving, and resume options.
Check the provided configs for examples.
Example Commands
- Pretraining a model:
python main.py --experiment PretrainExperiment --config configs/miccai2025/pretrain_stable.yaml - Training a task vector from a checkpoint:
python main.py --experiment TaskVectorTrainExperiment --config configs/miccai2025/finetune.yaml --override BackboneConfig.PRETRAIN_CHECKPOINTS="/path/to/checkpoint.pth"
For further details, refer to the config files used in our experiments under the configs folder.
5. Cite
If you used our work, please cite it:
@incollection{lumetti2025u,
title={U-Net Transplant: The Role of Pre-training for Model Merging in 3D Medical Segmentation},
author={Lumetti, Luca and Capitani, Giacomo and Ficarra, Elisa and Grana, Costantino and Calderara, Simone and Porrello, Angelo and Bolelli, Federico and others},
booktitle={Medical Image Computing and Computer Assisted Intervention--MICCAI 2025},
year={2025}
}