initial commit
Browse files- .gitattributes +1 -0
- README.md +3 -0
- cloud_mask_visualization.png +3 -0
- config.json +0 -0
- model.py +199 -0
- requirements.txt +8 -0
.gitattributes
CHANGED
|
@@ -33,3 +33,4 @@ saved_model/**/* filter=lfs diff=lfs merge=lfs -text
|
|
| 33 |
*.zip filter=lfs diff=lfs merge=lfs -text
|
| 34 |
*.zst filter=lfs diff=lfs merge=lfs -text
|
| 35 |
*tfevents* filter=lfs diff=lfs merge=lfs -text
|
|
|
|
|
|
| 33 |
*.zip filter=lfs diff=lfs merge=lfs -text
|
| 34 |
*.zst filter=lfs diff=lfs merge=lfs -text
|
| 35 |
*tfevents* filter=lfs diff=lfs merge=lfs -text
|
| 36 |
+
cloud_mask_visualization.png filter=lfs diff=lfs merge=lfs -text
|
README.md
CHANGED
|
@@ -1,3 +1,6 @@
|
|
| 1 |
---
|
| 2 |
license: mit
|
| 3 |
---
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
---
|
| 2 |
license: mit
|
| 3 |
---
|
| 4 |
+
|
| 5 |
+
|
| 6 |
+
Python 3.12
|
cloud_mask_visualization.png
ADDED
|
Git LFS Details
|
config.json
ADDED
|
File without changes
|
model.py
ADDED
|
@@ -0,0 +1,199 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
"""
|
| 2 |
+
Cloud Mask Prediction and Visualization Module
|
| 3 |
+
|
| 4 |
+
This script processes Sentinel-2 satellite imagery bands to predict cloud masks
|
| 5 |
+
using the omnicloudmask library. It reads blue, red, green, and near-infrared bands,
|
| 6 |
+
resamples them as needed, creates a stacked array for prediction, and visualizes
|
| 7 |
+
the cloud mask overlaid on the original RGB image.
|
| 8 |
+
"""
|
| 9 |
+
|
| 10 |
+
import rasterio
|
| 11 |
+
import numpy as np
|
| 12 |
+
from rasterio.enums import Resampling
|
| 13 |
+
from omnicloudmask import predict_from_array
|
| 14 |
+
import matplotlib.pyplot as plt
|
| 15 |
+
from matplotlib.colors import ListedColormap
|
| 16 |
+
import matplotlib.patches as mpatches
|
| 17 |
+
|
| 18 |
+
def load_band(file_path, resample=False, target_height=None, target_width=None):
|
| 19 |
+
"""
|
| 20 |
+
Load a single band from a raster file with optional resampling.
|
| 21 |
+
|
| 22 |
+
Args:
|
| 23 |
+
file_path (str): Path to the raster file
|
| 24 |
+
resample (bool): Whether to resample the band
|
| 25 |
+
target_height (int, optional): Target height for resampling
|
| 26 |
+
target_width (int, optional): Target width for resampling
|
| 27 |
+
|
| 28 |
+
Returns:
|
| 29 |
+
numpy.ndarray: Band data as float32 array
|
| 30 |
+
"""
|
| 31 |
+
with rasterio.open(file_path) as src:
|
| 32 |
+
if resample and target_height is not None and target_width is not None:
|
| 33 |
+
band_data = src.read(
|
| 34 |
+
out_shape=(src.count, target_height, target_width),
|
| 35 |
+
resampling=Resampling.bilinear
|
| 36 |
+
)[0].astype(np.float32)
|
| 37 |
+
else:
|
| 38 |
+
band_data = src.read()[0].astype(np.float32)
|
| 39 |
+
|
| 40 |
+
return band_data
|
| 41 |
+
|
| 42 |
+
def prepare_input_array(base_path="jp2s/"):
|
| 43 |
+
"""
|
| 44 |
+
Prepare a stacked array of satellite bands for cloud mask prediction.
|
| 45 |
+
|
| 46 |
+
This function loads blue, red, green, and near-infrared bands from Sentinel-2 imagery,
|
| 47 |
+
resamples the NIR band if needed (from 20m to 10m resolution), and stacks the required
|
| 48 |
+
bands for cloud mask prediction in CHW (channel, height, width) format.
|
| 49 |
+
|
| 50 |
+
Args:
|
| 51 |
+
base_path (str): Base directory containing the JP2 band files
|
| 52 |
+
|
| 53 |
+
Returns:
|
| 54 |
+
tuple: (stacked_array, rgb_image)
|
| 55 |
+
- stacked_array: numpy.ndarray with bands stacked in CHW format for prediction
|
| 56 |
+
- rgb_image: numpy.ndarray with RGB bands for visualization
|
| 57 |
+
"""
|
| 58 |
+
# Define paths to band files
|
| 59 |
+
band_paths = {
|
| 60 |
+
'blue': f"{base_path}B02.jp2", # Blue band (10m)
|
| 61 |
+
'green': f"{base_path}B03.jp2", # Green band (10m)
|
| 62 |
+
'red': f"{base_path}B04.jp2", # Red band (10m)
|
| 63 |
+
'nir': f"{base_path}B8A.jp2" # Near-infrared band (20m)
|
| 64 |
+
}
|
| 65 |
+
|
| 66 |
+
# Get dimensions from red band to use for resampling
|
| 67 |
+
with rasterio.open(band_paths['red']) as src:
|
| 68 |
+
target_height = src.height
|
| 69 |
+
target_width = src.width
|
| 70 |
+
|
| 71 |
+
# Load bands (resample NIR band to match 10m resolution)
|
| 72 |
+
blue_data = load_band(band_paths['blue'])
|
| 73 |
+
green_data = load_band(band_paths['green'])
|
| 74 |
+
red_data = load_band(band_paths['red'])
|
| 75 |
+
nir_data = load_band(
|
| 76 |
+
band_paths['nir'],
|
| 77 |
+
resample=True,
|
| 78 |
+
target_height=target_height,
|
| 79 |
+
target_width=target_width
|
| 80 |
+
)
|
| 81 |
+
|
| 82 |
+
# Print band shapes for debugging
|
| 83 |
+
print(f"Band shapes - Blue: {blue_data.shape}, Green: {green_data.shape}, Red: {red_data.shape}, NIR: {nir_data.shape}")
|
| 84 |
+
|
| 85 |
+
# Create RGB image for visualization (scale to 0-1 range)
|
| 86 |
+
# Adjust scaling factor based on your data's bit depth (e.g., 10000 for 16-bit Sentinel-2)
|
| 87 |
+
scale_factor = 10000.0 # Adjust based on your data
|
| 88 |
+
rgb_image = np.stack([
|
| 89 |
+
red_data / scale_factor,
|
| 90 |
+
green_data / scale_factor,
|
| 91 |
+
blue_data / scale_factor
|
| 92 |
+
], axis=-1)
|
| 93 |
+
|
| 94 |
+
# Clip values to 0-1 range
|
| 95 |
+
rgb_image = np.clip(rgb_image, 0, 1)
|
| 96 |
+
|
| 97 |
+
# Stack bands in CHW format for cloud mask prediction (red, green, nir)
|
| 98 |
+
prediction_array = np.stack([red_data, green_data, nir_data], axis=0)
|
| 99 |
+
|
| 100 |
+
return prediction_array, rgb_image
|
| 101 |
+
|
| 102 |
+
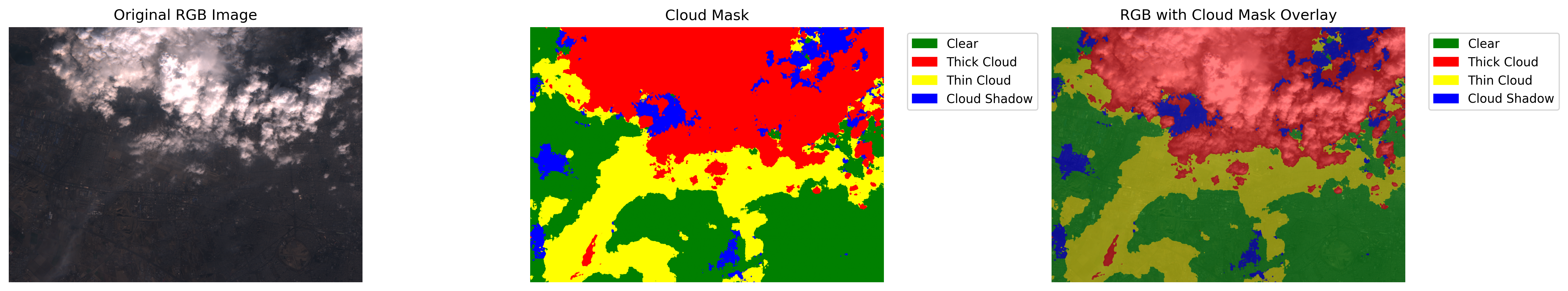
def visualize_cloud_mask(rgb_image, cloud_mask, output_path="cloud_mask_visualization.png"):
|
| 103 |
+
"""
|
| 104 |
+
Visualize the cloud mask overlaid on the original RGB image.
|
| 105 |
+
|
| 106 |
+
Args:
|
| 107 |
+
rgb_image (numpy.ndarray): RGB image array (HWC format)
|
| 108 |
+
cloud_mask (numpy.ndarray): Predicted cloud mask
|
| 109 |
+
output_path (str): Path to save the visualization
|
| 110 |
+
"""
|
| 111 |
+
# Fix the cloud mask shape if it has an extra dimension
|
| 112 |
+
if cloud_mask.ndim > 2:
|
| 113 |
+
# Check the shape and squeeze if needed
|
| 114 |
+
print(f"Original cloud mask shape: {cloud_mask.shape}")
|
| 115 |
+
cloud_mask = np.squeeze(cloud_mask)
|
| 116 |
+
print(f"Squeezed cloud mask shape: {cloud_mask.shape}")
|
| 117 |
+
|
| 118 |
+
# Create figure with two subplots
|
| 119 |
+
fig, (ax1, ax2, ax3) = plt.subplots(1, 3, figsize=(18, 6))
|
| 120 |
+
|
| 121 |
+
# Plot original RGB image
|
| 122 |
+
ax1.imshow(rgb_image)
|
| 123 |
+
ax1.set_title("Original RGB Image")
|
| 124 |
+
ax1.axis('off')
|
| 125 |
+
|
| 126 |
+
# Define colormap for cloud mask
|
| 127 |
+
# 0=Clear, 1=Thick Cloud, 2=Thin Cloud, 3=Cloud Shadow
|
| 128 |
+
cloud_cmap = ListedColormap(['green', 'red', 'yellow', 'blue'])
|
| 129 |
+
|
| 130 |
+
# Plot cloud mask
|
| 131 |
+
im = ax2.imshow(cloud_mask, cmap=cloud_cmap, vmin=0, vmax=3)
|
| 132 |
+
ax2.set_title("Cloud Mask")
|
| 133 |
+
ax2.axis('off')
|
| 134 |
+
|
| 135 |
+
# Create legend patches
|
| 136 |
+
legend_patches = [
|
| 137 |
+
mpatches.Patch(color='green', label='Clear'),
|
| 138 |
+
mpatches.Patch(color='red', label='Thick Cloud'),
|
| 139 |
+
mpatches.Patch(color='yellow', label='Thin Cloud'),
|
| 140 |
+
mpatches.Patch(color='blue', label='Cloud Shadow')
|
| 141 |
+
]
|
| 142 |
+
ax2.legend(handles=legend_patches, bbox_to_anchor=(1.05, 1), loc='upper left')
|
| 143 |
+
|
| 144 |
+
# Plot RGB with semi-transparent cloud mask overlay
|
| 145 |
+
ax3.imshow(rgb_image)
|
| 146 |
+
|
| 147 |
+
# Create a masked array with transparency
|
| 148 |
+
cloud_mask_rgba = np.zeros((*cloud_mask.shape, 4))
|
| 149 |
+
|
| 150 |
+
# Set colors with alpha for each class
|
| 151 |
+
cloud_mask_rgba[cloud_mask == 0] = [0, 1, 0, 0.3] # Clear - green with low opacity
|
| 152 |
+
cloud_mask_rgba[cloud_mask == 1] = [1, 0, 0, 0.5] # Thick Cloud - red
|
| 153 |
+
cloud_mask_rgba[cloud_mask == 2] = [1, 1, 0, 0.5] # Thin Cloud - yellow
|
| 154 |
+
cloud_mask_rgba[cloud_mask == 3] = [0, 0, 1, 0.5] # Cloud Shadow - blue
|
| 155 |
+
|
| 156 |
+
ax3.imshow(cloud_mask_rgba)
|
| 157 |
+
ax3.set_title("RGB with Cloud Mask Overlay")
|
| 158 |
+
ax3.axis('off')
|
| 159 |
+
|
| 160 |
+
# Add legend to the overlay plot as well
|
| 161 |
+
ax3.legend(handles=legend_patches, bbox_to_anchor=(1.05, 1), loc='upper left')
|
| 162 |
+
|
| 163 |
+
# Adjust layout and save
|
| 164 |
+
plt.tight_layout()
|
| 165 |
+
plt.savefig(output_path, dpi=300, bbox_inches='tight')
|
| 166 |
+
plt.show()
|
| 167 |
+
|
| 168 |
+
print(f"Visualization saved to {output_path}")
|
| 169 |
+
|
| 170 |
+
def main():
|
| 171 |
+
"""
|
| 172 |
+
Main function to run the cloud mask prediction and visualization workflow.
|
| 173 |
+
"""
|
| 174 |
+
# Create input array from satellite bands and get RGB image for visualization
|
| 175 |
+
input_array, rgb_image = prepare_input_array()
|
| 176 |
+
|
| 177 |
+
# Predict cloud mask using omnicloudmask
|
| 178 |
+
pred_mask = predict_from_array(input_array)
|
| 179 |
+
|
| 180 |
+
# Print prediction results and shape
|
| 181 |
+
print("Cloud mask prediction results:")
|
| 182 |
+
print(f"Cloud mask shape: {pred_mask.shape}")
|
| 183 |
+
print(f"Unique classes in mask: {np.unique(pred_mask)}")
|
| 184 |
+
|
| 185 |
+
# Calculate class distribution
|
| 186 |
+
if pred_mask.ndim > 2:
|
| 187 |
+
# Squeeze if needed for counting
|
| 188 |
+
flat_mask = np.squeeze(pred_mask)
|
| 189 |
+
else:
|
| 190 |
+
flat_mask = pred_mask
|
| 191 |
+
|
| 192 |
+
print(f"Class distribution: Clear: {np.sum(flat_mask == 0)}, Thick Cloud: {np.sum(flat_mask == 1)}, "
|
| 193 |
+
f"Thin Cloud: {np.sum(flat_mask == 2)}, Cloud Shadow: {np.sum(flat_mask == 3)}")
|
| 194 |
+
|
| 195 |
+
# Visualize the cloud mask on the original image
|
| 196 |
+
visualize_cloud_mask(rgb_image, pred_mask)
|
| 197 |
+
|
| 198 |
+
if __name__ == "__main__":
|
| 199 |
+
main()
|
requirements.txt
ADDED
|
@@ -0,0 +1,8 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
rasterio==1.3.11
|
| 2 |
+
matplotlib==3.7.5
|
| 3 |
+
fastai>=2.7
|
| 4 |
+
timm>=0.9
|
| 5 |
+
tqdm>=4.0
|
| 6 |
+
rasterio>=1.3
|
| 7 |
+
gdown>=5.1.0
|
| 8 |
+
torch>=2.2
|