Spaces:
Sleeping
Sleeping
Upload 20 files
Browse files- .gitattributes +2 -0
- ARTI_Dataset.csv +142 -0
- ARTI_Main_Data.csv +142 -0
- Output1.spv +3 -0
- Untitled9.sav +0 -0
- actual_vs_predicted_heatmap.png +0 -0
- app.py +65 -0
- class_distribution.png +0 -0
- confusion_matrix.png +0 -0
- confusion_matrix_multiclass.png +0 -0
- correlation_heatmap.png +3 -0
- feature_importance.png +0 -0
- feature_importance_rf.png +0 -0
- feature_label_encoders.pkl +3 -0
- main.py +146 -0
- mh.py +65 -0
- roc_curve.png +0 -0
- scaler.pkl +3 -0
- target_label_encoder.pkl +3 -0
- voting_model.pkl +3 -0
- voting_model_multiclass.pkl +3 -0
.gitattributes
CHANGED
|
@@ -33,3 +33,5 @@ saved_model/**/* filter=lfs diff=lfs merge=lfs -text
|
|
| 33 |
*.zip filter=lfs diff=lfs merge=lfs -text
|
| 34 |
*.zst filter=lfs diff=lfs merge=lfs -text
|
| 35 |
*tfevents* filter=lfs diff=lfs merge=lfs -text
|
|
|
|
|
|
|
|
|
| 33 |
*.zip filter=lfs diff=lfs merge=lfs -text
|
| 34 |
*.zst filter=lfs diff=lfs merge=lfs -text
|
| 35 |
*tfevents* filter=lfs diff=lfs merge=lfs -text
|
| 36 |
+
correlation_heatmap.png filter=lfs diff=lfs merge=lfs -text
|
| 37 |
+
Output1.spv filter=lfs diff=lfs merge=lfs -text
|
ARTI_Dataset.csv
ADDED
|
@@ -0,0 +1,142 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
Age,Sex,Socioeconomic_Status,Vitamin_D_Level_ng/ml,Vitamin_D_Status,Vitamin_D_Supplemented,ARTI_Severity,Bacterial_Infection,Viral_Infection,Co_Infection,IL6_pg/ml,IL8_pg/ml,Recovery_Days,Crowding_Index
|
| 2 |
+
13,Female,Middle,30,Insufficient,No,Pneumonia,Staphylococcus aureus,Adenovirus,No,75.53661948,74.92566999,10,2
|
| 3 |
+
4,Male,Low,40.6,Sufficient,No,Severe Pneumonia,Klebsiella pneumoniae,RSV,No,60.27978329,57.83197895,8,5
|
| 4 |
+
4,Male,Low,25.7,Insufficient,Yes,Severe Pneumonia,Staphylococcus aureus,RSV,Yes,67.60185751,66.58953992,4,3
|
| 5 |
+
10,Male,Middle,42.3,Sufficient,Yes,No Pneumonia,Staphylococcus aureus,Influenza A,No,58.44445114,62.95506693,4,3
|
| 6 |
+
8,Female,Low,12.2,Insufficient,No,Severe Pneumonia,Staphylococcus aureus,Adenovirus,Yes,82.49870863,94.13319318,8,1
|
| 7 |
+
6,Female,Low,40.9,Sufficient,Yes,No Pneumonia,Staphylococcus aureus,Influenza B,No,53.51027422,58.5649894,2,1
|
| 8 |
+
3,Female,High,16.2,Insufficient,No,Severe Pneumonia,Klebsiella pneumoniae,Rhinovirus,No,76.98648198,78.49099239,15,1
|
| 9 |
+
10,Female,Middle,17.2,Insufficient,No,Pneumonia,Staphylococcus aureus,Adenovirus,Yes,73.2437316,100.328528,14,4
|
| 10 |
+
5,Female,Middle,18.8,Insufficient,Yes,Severe Pneumonia,Klebsiella pneumoniae,RSV,Yes,78.36418416,83.92054104,2,3
|
| 11 |
+
6,Female,Middle,17.8,Insufficient,Yes,Severe Pneumonia,Klebsiella pneumoniae,RSV,Yes,87.84444372,72.00744812,2,3
|
| 12 |
+
5,Female,Middle,20.9,Insufficient,Yes,Pneumonia,None,None,No,90.38242008,90.41232369,3,1
|
| 13 |
+
6,Male,Low,14.2,Insufficient,No,Pneumonia,Klebsiella pneumoniae,RSV,Yes,82.93207421,78.953802,14,5
|
| 14 |
+
8,Female,Low,28.3,Insufficient,No,No Pneumonia,Klebsiella pneumoniae,Adenovirus,Yes,77.13052763,93.63502672,14,4
|
| 15 |
+
7,Male,Low,14.5,Insufficient,Yes,Pneumonia,Streptococcus pneumoniae,Influenza B,No,83.52708841,86.13726286,2,2
|
| 16 |
+
5,Male,Low,26.6,Insufficient,Yes,Pneumonia,Staphylococcus aureus,Adenovirus,Yes,65.1463214,81.50652829,4,1
|
| 17 |
+
10,Female,Middle,35.3,Sufficient,Yes,Severe Pneumonia,Klebsiella pneumoniae,RSV,Yes,68.20471131,48.05985951,4,1
|
| 18 |
+
6,Female,Low,16.4,Insufficient,No,Severe Pneumonia,Staphylococcus aureus,RSV,Yes,84.77407043,73.32687293,14,2
|
| 19 |
+
4,Male,Low,27.9,Insufficient,Yes,Pneumonia,Klebsiella pneumoniae,RSV,Yes,81.19009804,82.77087696,2,3
|
| 20 |
+
12,Male,Low,25.6,Insufficient,No,No Pneumonia,Klebsiella pneumoniae,RSV,Yes,66.72660759,76.94252053,12,2
|
| 21 |
+
9,Female,Low,42.5,Sufficient,Yes,Severe Pneumonia,Staphylococcus aureus,Adenovirus,Yes,71.48016354,73.83917138,2,2
|
| 22 |
+
13,Female,Low,15,Insufficient,Yes,No Pneumonia,Klebsiella pneumoniae,RSV,Yes,92.95858662,88.88140084,5,3
|
| 23 |
+
4,Male,Low,28,Insufficient,No,Pneumonia,Staphylococcus aureus,RSV,Yes,71.91038213,73.16236992,12,1
|
| 24 |
+
1,Female,Low,5,Deficient,Yes,Pneumonia,Klebsiella pneumoniae,Influenza B,No,93.50681104,108.7085919,4,2
|
| 25 |
+
7,Female,Low,15,Insufficient,No,Pneumonia,Klebsiella pneumoniae,RSV,No,81.42095066,93.08109599,13,2
|
| 26 |
+
7,Female,Low,23.5,Insufficient,No,No Pneumonia,Klebsiella pneumoniae,Adenovirus,Yes,77.0486632,74.92991041,9,2
|
| 27 |
+
4,Male,Middle,33.9,Sufficient,Yes,Pneumonia,None,Parainfluenza,No,65.18031922,57.3471672,3,3
|
| 28 |
+
13,Male,Low,7.7,Deficient,Yes,No Pneumonia,Klebsiella pneumoniae,RSV,Yes,94.86897707,73.32707468,5,1
|
| 29 |
+
3,Male,Low,27.9,Insufficient,Yes,Pneumonia,Klebsiella pneumoniae,RSV,No,70.62816669,68.59867464,2,3
|
| 30 |
+
8,Female,Middle,19.4,Insufficient,Yes,Pneumonia,Klebsiella pneumoniae,Parainfluenza,No,87.67481846,71.10931924,2,3
|
| 31 |
+
5,Female,Low,5,Deficient,No,No Pneumonia,Staphylococcus aureus,RSV,Yes,95.62523016,115.7816371,10,1
|
| 32 |
+
10,Male,High,21.8,Insufficient,No,Severe Pneumonia,Klebsiella pneumoniae,Rhinovirus,No,76.91591486,72.99703245,10,3
|
| 33 |
+
7,Male,Middle,37.1,Sufficient,No,Severe Pneumonia,Klebsiella pneumoniae,RSV,Yes,68.02143291,57.47832795,14,1
|
| 34 |
+
4,Male,Low,8.8,Deficient,Yes,Pneumonia,Klebsiella pneumoniae,RSV,Yes,83.08738185,79.030041,4,1
|
| 35 |
+
4,Female,Middle,18.9,Insufficient,No,Severe Pneumonia,Klebsiella pneumoniae,Adenovirus,Yes,82.80494176,78.96946137,13,4
|
| 36 |
+
8,Male,Low,10.1,Insufficient,Yes,No Pneumonia,Klebsiella pneumoniae,RSV,Yes,90.02785668,84.58372056,2,5
|
| 37 |
+
8,Male,Middle,15.3,Insufficient,Yes,No Pneumonia,Klebsiella pneumoniae,RSV,Yes,77.64574735,98.60183543,3,4
|
| 38 |
+
2,Female,Low,34.3,Sufficient,No,Severe Pneumonia,Staphylococcus aureus,Adenovirus,Yes,57.1574836,67.04780928,15,2
|
| 39 |
+
2,Female,High,17.3,Insufficient,Yes,Severe Pneumonia,Staphylococcus aureus,Influenza B,No,87.75172501,82.62908782,4,1
|
| 40 |
+
3,Female,Low,34,Sufficient,No,No Pneumonia,Klebsiella pneumoniae,RSV,Yes,70.81395345,64.92261797,11,3
|
| 41 |
+
12,Male,Middle,31.1,Sufficient,No,No Pneumonia,None,Adenovirus,No,69.76279215,75.18939936,9,2
|
| 42 |
+
12,Female,Middle,16.4,Insufficient,Yes,Pneumonia,Staphylococcus aureus,Parainfluenza,No,92.18328888,79.98137467,2,1
|
| 43 |
+
3,Female,Low,15,Insufficient,No,Severe Pneumonia,Klebsiella pneumoniae,RSV,No,87.20484091,90.33764875,13,2
|
| 44 |
+
1,Male,Low,11,Insufficient,No,No Pneumonia,Klebsiella pneumoniae,RSV,Yes,95.2490266,101.9117586,9,1
|
| 45 |
+
3,Female,Middle,10.7,Insufficient,Yes,Pneumonia,Klebsiella pneumoniae,RSV,Yes,91.40162294,92.98060008,4,4
|
| 46 |
+
6,Male,High,23.5,Insufficient,No,Severe Pneumonia,Staphylococcus aureus,Rhinovirus,No,75.12095217,74.50214974,9,2
|
| 47 |
+
8,Male,Low,19.8,Insufficient,No,Severe Pneumonia,Staphylococcus aureus,RSV,No,74.91234374,72.57232663,9,3
|
| 48 |
+
13,Female,Middle,13.8,Insufficient,Yes,Severe Pneumonia,Staphylococcus aureus,RSV,Yes,83.18281873,83.99900811,2,2
|
| 49 |
+
12,Female,High,5,Deficient,Yes,Pneumonia,Streptococcus pneumoniae,Influenza B,No,93.71438864,99.50463099,2,3
|
| 50 |
+
10,Female,High,21.1,Insufficient,No,Pneumonia,Staphylococcus aureus,Adenovirus,No,85.71576551,66.63978126,13,3
|
| 51 |
+
6,Female,Low,15.3,Insufficient,Yes,Severe Pneumonia,Staphylococcus aureus,RSV,Yes,88.54959779,91.35234206,2,1
|
| 52 |
+
5,Female,Middle,18,Insufficient,No,Pneumonia,Staphylococcus aureus,Influenza A,No,83.65159534,83.66090151,14,5
|
| 53 |
+
4,Female,Low,23,Insufficient,Yes,No Pneumonia,Streptococcus pneumoniae,RSV,No,81.35004954,75.80648672,3,3
|
| 54 |
+
3,Male,High,5,Deficient,No,No Pneumonia,Klebsiella pneumoniae,RSV,Yes,97.63691858,89.72469271,10,4
|
| 55 |
+
13,Male,Low,5,Deficient,No,No Pneumonia,Klebsiella pneumoniae,RSV,Yes,96.30968806,91.18897839,10,4
|
| 56 |
+
4,Female,Low,43,Sufficient,Yes,No Pneumonia,Staphylococcus aureus,RSV,Yes,49.50670299,61.77844833,3,3
|
| 57 |
+
7,Male,Middle,14.9,Insufficient,Yes,No Pneumonia,Staphylococcus aureus,Adenovirus,Yes,90.79578786,84.07430779,4,4
|
| 58 |
+
1,Female,Low,5,Deficient,No,Pneumonia,Klebsiella pneumoniae,RSV,Yes,92.70744076,90.99088026,14,1
|
| 59 |
+
7,Male,High,5.5,Deficient,Yes,Pneumonia,Streptococcus pneumoniae,None,No,99.94270139,98.24850795,2,1
|
| 60 |
+
1,Male,High,44.8,Sufficient,No,Pneumonia,Staphylococcus aureus,RSV,No,61.12605146,59.26551822,9,1
|
| 61 |
+
2,Male,Middle,13,Insufficient,Yes,No Pneumonia,Streptococcus pneumoniae,Influenza B,No,83.64980403,77.00659951,2,2
|
| 62 |
+
4,Female,Low,31.6,Sufficient,No,Pneumonia,Staphylococcus aureus,Parainfluenza,No,73.1485804,72.97184557,11,2
|
| 63 |
+
1,Male,Middle,21.2,Insufficient,No,Pneumonia,Streptococcus pneumoniae,None,No,76.9950626,98.8135857,8,2
|
| 64 |
+
11,Female,Low,15.2,Insufficient,No,Severe Pneumonia,Streptococcus pneumoniae,Influenza A,No,83.02483836,88.55696632,13,4
|
| 65 |
+
12,Male,Low,27.8,Insufficient,Yes,No Pneumonia,Klebsiella pneumoniae,RSV,Yes,73.48736617,58.60794236,3,2
|
| 66 |
+
11,Male,High,5,Deficient,Yes,Pneumonia,Staphylococcus aureus,Metapneumovirus,No,96.42236295,96.41900313,2,2
|
| 67 |
+
11,Male,Low,5.6,Deficient,Yes,No Pneumonia,Streptococcus pneumoniae,Influenza B,No,94.59275758,79.75347701,3,3
|
| 68 |
+
9,Male,Low,15.1,Insufficient,Yes,Pneumonia,Klebsiella pneumoniae,Adenovirus,Yes,78.22855992,110.2389734,2,1
|
| 69 |
+
12,Male,High,6.6,Deficient,Yes,Pneumonia,Staphylococcus aureus,RSV,No,92.7927668,111.2853975,4,4
|
| 70 |
+
6,Female,Low,14.2,Insufficient,Yes,Severe Pneumonia,None,Adenovirus,No,97.19239364,80.47249214,2,1
|
| 71 |
+
13,Female,Low,28.5,Insufficient,No,Pneumonia,Klebsiella pneumoniae,RSV,Yes,57.57997886,53.53466241,8,5
|
| 72 |
+
1,Male,High,21.3,Insufficient,No,No Pneumonia,Klebsiella pneumoniae,RSV,No,83.0487022,71.6730305,15,3
|
| 73 |
+
1,Male,Low,33.3,Sufficient,Yes,Severe Pneumonia,Staphylococcus aureus,RSV,Yes,71.98562467,65.25207409,2,1
|
| 74 |
+
6,Male,Middle,22,Insufficient,No,Pneumonia,None,Influenza A,No,71.2082614,63.90509006,14,1
|
| 75 |
+
4,Male,High,29.9,Insufficient,No,No Pneumonia,Staphylococcus aureus,RSV,Yes,67.75280543,69.54781988,11,4
|
| 76 |
+
4,Female,High,19.4,Insufficient,Yes,No Pneumonia,Klebsiella pneumoniae,RSV,Yes,75.59641421,79.09937069,5,3
|
| 77 |
+
11,Male,Middle,14.1,Insufficient,No,Pneumonia,None,Adenovirus,No,95.16982689,86.90123758,13,3
|
| 78 |
+
6,Female,Low,34.8,Sufficient,No,No Pneumonia,Klebsiella pneumoniae,RSV,Yes,70.71512633,67.03716142,14,3
|
| 79 |
+
12,Male,Low,12.9,Insufficient,No,Pneumonia,None,Influenza A,No,92.85712687,79.3750592,9,1
|
| 80 |
+
11,Female,High,23.4,Insufficient,Yes,Severe Pneumonia,Klebsiella pneumoniae,None,No,77.20323613,69.77268768,3,2
|
| 81 |
+
7,Male,High,27.7,Insufficient,No,Pneumonia,Klebsiella pneumoniae,RSV,No,78.86045975,72.66473732,10,1
|
| 82 |
+
9,Male,Middle,28.4,Insufficient,No,Severe Pneumonia,Klebsiella pneumoniae,RSV,Yes,71.55881716,66.55206296,8,4
|
| 83 |
+
10,Female,Low,8.3,Deficient,Yes,No Pneumonia,Klebsiella pneumoniae,RSV,Yes,98.49578814,98.95995815,2,3
|
| 84 |
+
5,Male,Low,30.5,Sufficient,No,Severe Pneumonia,Klebsiella pneumoniae,Adenovirus,Yes,68.75651788,69.7871564,12,2
|
| 85 |
+
9,Male,Low,10.2,Insufficient,No,No Pneumonia,Klebsiella pneumoniae,Adenovirus,Yes,86.40379042,104.7200259,12,1
|
| 86 |
+
4,Male,Middle,28.1,Insufficient,No,Severe Pneumonia,Klebsiella pneumoniae,RSV,Yes,80.95475133,78.46385951,11,2
|
| 87 |
+
13,Female,Low,25.4,Insufficient,No,Severe Pneumonia,Klebsiella pneumoniae,RSV,Yes,74.4957562,83.42971096,9,2
|
| 88 |
+
10,Male,High,28.2,Insufficient,No,No Pneumonia,Staphylococcus aureus,RSV,No,67.30600046,72.86052854,15,3
|
| 89 |
+
2,Male,Middle,7.9,Deficient,No,Pneumonia,Staphylococcus aureus,RSV,Yes,87.55183051,93.35889201,8,3
|
| 90 |
+
3,Female,Low,25.9,Insufficient,Yes,Severe Pneumonia,Staphylococcus aureus,Adenovirus,Yes,69.56710663,87.95056464,2,4
|
| 91 |
+
13,Female,Middle,30.9,Sufficient,No,No Pneumonia,Staphylococcus aureus,Parainfluenza,No,73.32221176,66.07365693,9,2
|
| 92 |
+
11,Female,Low,21.9,Insufficient,No,Pneumonia,Klebsiella pneumoniae,Adenovirus,Yes,77.60568561,82.47423483,8,1
|
| 93 |
+
1,Male,Middle,11.8,Insufficient,No,Pneumonia,Streptococcus pneumoniae,Rhinovirus,No,99.68979488,86.07009974,11,3
|
| 94 |
+
6,Male,Middle,29.2,Insufficient,No,No Pneumonia,Staphylococcus aureus,Influenza A,No,76.73089091,68.65091153,11,3
|
| 95 |
+
13,Male,High,21.1,Insufficient,No,No Pneumonia,Staphylococcus aureus,RSV,No,80.18569065,69.54882987,15,3
|
| 96 |
+
6,Female,High,5,Deficient,Yes,Severe Pneumonia,None,None,No,102.0194877,89.05988519,5,3
|
| 97 |
+
1,Male,Low,11,Insufficient,No,Severe Pneumonia,Klebsiella pneumoniae,None,No,86.52246332,81.57384439,12,2
|
| 98 |
+
11,Male,Low,14.7,Insufficient,No,Pneumonia,Klebsiella pneumoniae,RSV,Yes,76.02607098,90.05495047,9,3
|
| 99 |
+
13,Female,Middle,35.1,Sufficient,No,No Pneumonia,Staphylococcus aureus,Adenovirus,No,59.95208471,73.22164743,13,2
|
| 100 |
+
3,Female,Low,33.3,Sufficient,Yes,Severe Pneumonia,Klebsiella pneumoniae,Adenovirus,Yes,67.43603915,65.94197301,5,1
|
| 101 |
+
5,Female,Low,33.3,Sufficient,Yes,Pneumonia,Klebsiella pneumoniae,Influenza B,No,65.58729264,68.53702344,3,1
|
| 102 |
+
3,Female,Low,22.3,Insufficient,No,No Pneumonia,Staphylococcus aureus,RSV,No,81.82207694,76.48022916,12,1
|
| 103 |
+
10,Female,Middle,29.1,Insufficient,Yes,No Pneumonia,Klebsiella pneumoniae,Metapneumovirus,No,70.27693109,64.41379264,5,3
|
| 104 |
+
3,Male,Low,8.6,Deficient,Yes,No Pneumonia,None,Adenovirus,No,95.14017363,98.2981042,4,2
|
| 105 |
+
10,Female,Middle,20.1,Insufficient,Yes,No Pneumonia,Klebsiella pneumoniae,Adenovirus,No,87.40485815,85.12027755,5,3
|
| 106 |
+
3,Male,High,24.5,Insufficient,Yes,Severe Pneumonia,Klebsiella pneumoniae,RSV,No,74.86152075,60.35963212,4,4
|
| 107 |
+
5,Male,Low,9.3,Deficient,Yes,No Pneumonia,Klebsiella pneumoniae,RSV,Yes,93.47572644,88.43512799,3,2
|
| 108 |
+
13,Female,Low,22.1,Insufficient,No,Severe Pneumonia,Streptococcus pneumoniae,None,No,69.1260658,70.84378127,9,2
|
| 109 |
+
5,Female,Low,36.9,Sufficient,Yes,Severe Pneumonia,None,Metapneumovirus,No,62.48053011,59.97249144,3,3
|
| 110 |
+
4,Female,High,25.6,Insufficient,No,Severe Pneumonia,Streptococcus pneumoniae,Metapneumovirus,Yes,68.24020937,69.03654746,12,2
|
| 111 |
+
1,Male,Low,29.2,Insufficient,No,No Pneumonia,Staphylococcus aureus,RSV,No,71.52613389,70.42863703,13,1
|
| 112 |
+
5,Female,Low,5,Deficient,No,Severe Pneumonia,Klebsiella pneumoniae,RSV,Yes,96.17999878,92.36068212,10,2
|
| 113 |
+
10,Female,Low,35.6,Sufficient,Yes,No Pneumonia,Klebsiella pneumoniae,RSV,Yes,65.56338179,61.59087445,3,1
|
| 114 |
+
5,Female,Middle,18.1,Insufficient,No,Pneumonia,Klebsiella pneumoniae,RSV,Yes,79.49938705,90.75910743,10,4
|
| 115 |
+
4,Female,Low,12.3,Insufficient,No,No Pneumonia,Staphylococcus aureus,RSV,Yes,99.19234431,92.15742929,15,4
|
| 116 |
+
6,Female,Middle,18.3,Insufficient,No,No Pneumonia,Streptococcus pneumoniae,Influenza B,No,75.35989244,76.0688428,10,3
|
| 117 |
+
10,Male,High,5.1,Deficient,Yes,Severe Pneumonia,Klebsiella pneumoniae,Adenovirus,Yes,88.25427086,90.45562614,3,4
|
| 118 |
+
4,Male,Low,30.5,Sufficient,No,Pneumonia,Klebsiella pneumoniae,RSV,No,77.77489188,73.43434444,10,4
|
| 119 |
+
7,Male,High,16.6,Insufficient,Yes,No Pneumonia,None,RSV,No,79.87860435,87.32335657,3,1
|
| 120 |
+
7,Female,Low,12.6,Insufficient,No,Severe Pneumonia,Klebsiella pneumoniae,RSV,Yes,82.67654656,82.56975809,14,4
|
| 121 |
+
2,Female,High,18.3,Insufficient,No,No Pneumonia,Klebsiella pneumoniae,Metapneumovirus,No,93.21455949,79.82807466,8,3
|
| 122 |
+
3,Female,Middle,31.2,Sufficient,No,No Pneumonia,Staphylococcus aureus,RSV,Yes,73.72861358,83.57444832,14,4
|
| 123 |
+
1,Male,High,21.3,Insufficient,No,Severe Pneumonia,Staphylococcus aureus,Rhinovirus,No,75.72767726,74.00761529,14,4
|
| 124 |
+
8,Female,Low,5.9,Deficient,Yes,Pneumonia,Klebsiella pneumoniae,Adenovirus,Yes,95.18753953,109.3864367,4,2
|
| 125 |
+
9,Female,Low,24.6,Insufficient,No,Severe Pneumonia,Klebsiella pneumoniae,RSV,Yes,85.12935642,84.22933554,15,1
|
| 126 |
+
4,Female,Low,10.7,Insufficient,Yes,Severe Pneumonia,Staphylococcus aureus,RSV,No,92.7581034,103.1033302,4,2
|
| 127 |
+
6,Male,Low,5,Deficient,Yes,Pneumonia,Klebsiella pneumoniae,RSV,Yes,101.4307423,89.96742422,3,2
|
| 128 |
+
9,Female,Low,35,Sufficient,Yes,No Pneumonia,Klebsiella pneumoniae,Metapneumovirus,No,69.02401589,63.65155301,5,4
|
| 129 |
+
6,Male,Low,24.6,Insufficient,Yes,Severe Pneumonia,Klebsiella pneumoniae,RSV,Yes,80.05126673,71.80668953,2,4
|
| 130 |
+
12,Female,Low,19.2,Insufficient,No,Pneumonia,Staphylococcus aureus,RSV,Yes,80.99870848,89.81402554,14,1
|
| 131 |
+
12,Male,Low,16.5,Insufficient,Yes,Pneumonia,Klebsiella pneumoniae,RSV,Yes,83.55674239,78.95852982,3,3
|
| 132 |
+
12,Male,High,34,Sufficient,Yes,Severe Pneumonia,Klebsiella pneumoniae,RSV,Yes,67.33525193,71.84972395,5,2
|
| 133 |
+
9,Male,Low,50.9,Sufficient,No,No Pneumonia,Streptococcus pneumoniae,Influenza A,No,52.77226404,54.52639529,10,2
|
| 134 |
+
4,Female,Low,24.5,Insufficient,No,Pneumonia,Staphylococcus aureus,Adenovirus,Yes,74.08342342,75.11576375,13,1
|
| 135 |
+
8,Female,Low,9.7,Deficient,No,Pneumonia,Staphylococcus aureus,RSV,No,90.91026967,100.6892779,15,2
|
| 136 |
+
9,Male,Low,18.5,Insufficient,No,Severe Pneumonia,Klebsiella pneumoniae,RSV,Yes,79.94827192,82.36143896,15,4
|
| 137 |
+
4,Male,High,24.3,Insufficient,No,Pneumonia,Streptococcus pneumoniae,Metapneumovirus,Yes,76.27842411,82.37268575,11,2
|
| 138 |
+
5,Female,Low,5,Deficient,Yes,Severe Pneumonia,Staphylococcus aureus,Adenovirus,No,88.74501835,91.04237305,2,3
|
| 139 |
+
8,Female,Middle,23.3,Insufficient,Yes,Severe Pneumonia,Staphylococcus aureus,Adenovirus,Yes,89.72988733,70.91250549,3,3
|
| 140 |
+
12,Male,High,24.4,Insufficient,No,No Pneumonia,Streptococcus pneumoniae,RSV,No,71.93529941,81.44916423,15,2
|
| 141 |
+
10,Female,High,33.3,Sufficient,Yes,Severe Pneumonia,Staphylococcus aureus,Adenovirus,Yes,64.89269638,70.14672195,3,5
|
| 142 |
+
6,Female,Middle,32.2,Sufficient,No,Pneumonia,Staphylococcus aureus,RSV,Yes,63.70367803,69.74646131,11,5
|
ARTI_Main_Data.csv
ADDED
|
@@ -0,0 +1,142 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
Age,Sex,Socioeconomic_Status,Vitamin_D_Level_ng/ml,Vitamin_D_Status,Vitamin_D_Supplemented,ARTI_Severity,Bacterial_Infection,Viral_Infection,Co_Infection,IL6_pg/ml,IL8_pg/ml,Recovery_Days,Crowding_Index
|
| 2 |
+
13,Female,Middle,30.0,Insufficient,No,Pneumonia,Staphylococcus aureus,Adenovirus,No,75.53661948,74.92566999,10,2
|
| 3 |
+
4,Male,Low,40.6,Sufficient,No,Severe Pneumonia,Staphylococcus aureus,RSV,Yes,60.27978329,57.83197895,8,5
|
| 4 |
+
4,Male,Middle,25.7,Insufficient,Yes,Severe Pneumonia,Staphylococcus aureus,RSV,No,67.60185751,66.58953992,4,5
|
| 5 |
+
10,Male,Middle,42.3,Sufficient,Yes,No Pneumonia,Staphylococcus aureus,Influenza A,Yes,58.44445114,62.95506693,4,1
|
| 6 |
+
8,Female,Low,12.2,Insufficient,No,Severe Pneumonia,Klebsiella pneumoniae,RSV,No,82.49870863,94.13319318,8,5
|
| 7 |
+
6,Female,Middle,40.9,Sufficient,Yes,No Pneumonia,Staphylococcus aureus,Influenza B,No,53.51027422,58.5649894,2,2
|
| 8 |
+
3,Female,Low,16.2,Insufficient,No,Severe Pneumonia,Staphylococcus aureus,RSV,Yes,76.98648198,78.49099239,15,5
|
| 9 |
+
10,Female,Middle,17.2,Insufficient,No,Pneumonia,Staphylococcus aureus,Adenovirus,Yes,73.2437316,100.328528,14,4
|
| 10 |
+
5,Female,Low,18.8,Insufficient,Yes,Severe Pneumonia,Staphylococcus aureus,RSV,Yes,78.36418416,83.92054104,2,4
|
| 11 |
+
6,Female,Middle,17.8,Insufficient,Yes,Severe Pneumonia,Staphylococcus aureus,RSV,Yes,87.84444372,72.00744812,2,6
|
| 12 |
+
5,Female,Middle,20.9,Insufficient,Yes,Pneumonia,None,None,No,90.38242008,90.41232369,3,1
|
| 13 |
+
6,Male,Low,14.2,Insufficient,No,Pneumonia,Klebsiella pneumoniae,RSV,Yes,82.93207421,78.953802,14,5
|
| 14 |
+
8,Female,Middle,28.3,Insufficient,No,No Pneumonia,Klebsiella pneumoniae,Adenovirus,No,77.13052763,93.63502672,14,1
|
| 15 |
+
7,Male,Low,14.5,Insufficient,Yes,Pneumonia,Streptococcus pneumoniae,Influenza B,No,83.52708841,86.13726286,2,2
|
| 16 |
+
5,Male,Low,26.6,Insufficient,Yes,Pneumonia,Staphylococcus aureus,Adenovirus,Yes,65.1463214,81.50652829,4,1
|
| 17 |
+
10,Female,Low,35.3,Sufficient,Yes,Severe Pneumonia,Staphylococcus aureus,RSV,Yes,68.20471131,48.05985951,4,5
|
| 18 |
+
6,Female,Low,16.4,Insufficient,No,Severe Pneumonia,Klebsiella pneumoniae,RSV,Yes,84.77407043,73.32687293,14,5
|
| 19 |
+
4,Male,Low,27.9,Insufficient,Yes,Pneumonia,Klebsiella pneumoniae,RSV,Yes,81.19009804,82.77087696,2,3
|
| 20 |
+
12,Male,Middle,25.6,Insufficient,No,No Pneumonia,Klebsiella pneumoniae,RSV,No,66.72660759,76.94252053,12,1
|
| 21 |
+
9,Female,Low,42.5,Sufficient,Yes,Severe Pneumonia,Klebsiella pneumoniae,RSV,Yes,71.48016354,73.83917138,2,5
|
| 22 |
+
13,Female,Middle,15.0,Insufficient,Yes,No Pneumonia,Klebsiella pneumoniae,RSV,No,92.95858662,88.88140084,5,2
|
| 23 |
+
4,Male,Low,28.0,Insufficient,No,Pneumonia,Staphylococcus aureus,RSV,Yes,71.91038213,73.16236992,12,1
|
| 24 |
+
1,Female,Low,5.0,Deficient,Yes,Pneumonia,Klebsiella pneumoniae,Influenza B,No,93.50681104,108.7085919,4,2
|
| 25 |
+
7,Female,Low,15.0,Insufficient,No,Pneumonia,Klebsiella pneumoniae,RSV,No,81.42095066,93.08109599,13,2
|
| 26 |
+
7,Female,Middle,23.5,Insufficient,No,No Pneumonia,Klebsiella pneumoniae,Adenovirus,No,77.0486632,74.92991041,9,2
|
| 27 |
+
4,Male,Middle,33.9,Sufficient,Yes,Pneumonia,None,Parainfluenza,No,65.18031922,57.3471672,3,3
|
| 28 |
+
13,Male,High,7.7,Deficient,Yes,No Pneumonia,Klebsiella pneumoniae,RSV,Yes,94.86897707,73.32707468,5,2
|
| 29 |
+
3,Male,Low,27.9,Insufficient,Yes,Pneumonia,Klebsiella pneumoniae,RSV,No,70.62816669,68.59867464,2,3
|
| 30 |
+
8,Female,Middle,19.4,Insufficient,Yes,Pneumonia,Klebsiella pneumoniae,Parainfluenza,No,87.67481846,71.10931924,2,3
|
| 31 |
+
5,Female,Middle,5.0,Deficient,No,No Pneumonia,Staphylococcus aureus,RSV,No,95.62523016,115.7816371,10,1
|
| 32 |
+
10,Male,Low,21.8,Insufficient,No,Severe Pneumonia,Klebsiella pneumoniae,RSV,Yes,76.91591486,72.99703245,10,5
|
| 33 |
+
7,Male,Low,37.1,Sufficient,No,Severe Pneumonia,Staphylococcus aureus,Influenza A,Yes,68.02143291,57.47832795,14,5
|
| 34 |
+
4,Male,Low,8.8,Deficient,Yes,Pneumonia,Klebsiella pneumoniae,RSV,Yes,83.08738185,79.030041,4,1
|
| 35 |
+
4,Female,Low,18.9,Insufficient,No,Severe Pneumonia,Staphylococcus aureus,RSV,No,82.80494176,78.96946137,13,5
|
| 36 |
+
8,Male,High,10.1,Insufficient,Yes,No Pneumonia,Klebsiella pneumoniae,RSV,No,90.02785668,84.58372056,2,2
|
| 37 |
+
8,Male,High,15.3,Insufficient,Yes,No Pneumonia,Klebsiella pneumoniae,RSV,No,77.64574735,98.60183543,3,1
|
| 38 |
+
2,Female,Low,34.3,Sufficient,No,Severe Pneumonia,Klebsiella pneumoniae,Influenza A,Yes,57.1574836,67.04780928,15,6
|
| 39 |
+
2,Female,Low,17.3,Insufficient,Yes,Severe Pneumonia,Klebsiella pneumoniae,RSV,No,87.75172501,82.62908782,4,6
|
| 40 |
+
3,Female,Middle,34.0,Sufficient,No,No Pneumonia,Klebsiella pneumoniae,RSV,No,70.81395345,64.92261797,11,1
|
| 41 |
+
12,Male,Middle,31.1,Sufficient,No,No Pneumonia,None,Adenovirus,No,69.76279215,75.18939936,9,2
|
| 42 |
+
12,Female,Middle,16.4,Insufficient,Yes,Pneumonia,Staphylococcus aureus,Parainfluenza,No,92.18328888,79.98137467,2,1
|
| 43 |
+
3,Female,Middle,15.0,Insufficient,No,Severe Pneumonia,Klebsiella pneumoniae,Influenza A,Yes,87.20484091,90.33764875,13,5
|
| 44 |
+
1,Male,Middle,11.0,Insufficient,No,No Pneumonia,Klebsiella pneumoniae,RSV,No,95.2490266,101.9117586,9,2
|
| 45 |
+
3,Female,Middle,10.7,Insufficient,Yes,Pneumonia,Klebsiella pneumoniae,RSV,Yes,91.40162294,92.98060008,4,4
|
| 46 |
+
6,Male,Low,23.5,Insufficient,No,Severe Pneumonia,Klebsiella pneumoniae,Influenza A,Yes,75.12095217,74.50214974,9,6
|
| 47 |
+
8,Male,Middle,19.8,Insufficient,No,Severe Pneumonia,Staphylococcus aureus,RSV,Yes,74.91234374,72.57232663,9,4
|
| 48 |
+
13,Female,Middle,13.8,Insufficient,Yes,Severe Pneumonia,Klebsiella pneumoniae,RSV,Yes,83.18281873,83.99900811,2,5
|
| 49 |
+
12,Female,High,5.0,Deficient,Yes,Pneumonia,Streptococcus pneumoniae,Influenza B,No,93.71438864,99.50463099,2,3
|
| 50 |
+
10,Female,High,21.1,Insufficient,No,Pneumonia,Staphylococcus aureus,Adenovirus,No,85.71576551,66.63978126,13,3
|
| 51 |
+
6,Female,Middle,15.3,Insufficient,Yes,Severe Pneumonia,Klebsiella pneumoniae,RSV,No,88.54959779,91.35234206,2,4
|
| 52 |
+
5,Female,Middle,18.0,Insufficient,No,Pneumonia,Staphylococcus aureus,Influenza A,No,83.65159534,83.66090151,14,5
|
| 53 |
+
4,Female,Middle,23.0,Insufficient,Yes,No Pneumonia,Streptococcus pneumoniae,RSV,No,81.35004954,75.80648672,3,1
|
| 54 |
+
3,Male,Middle,5.0,Deficient,No,No Pneumonia,Klebsiella pneumoniae,RSV,Yes,97.63691858,89.72469271,10,1
|
| 55 |
+
13,Male,Middle,5.0,Deficient,No,No Pneumonia,Klebsiella pneumoniae,RSV,Yes,96.30968806,91.18897839,10,2
|
| 56 |
+
4,Female,Middle,43.0,Sufficient,Yes,No Pneumonia,Staphylococcus aureus,RSV,No,49.50670299,61.77844833,3,2
|
| 57 |
+
7,Male,Middle,14.9,Insufficient,Yes,No Pneumonia,Staphylococcus aureus,Adenovirus,No,90.79578786,84.07430779,4,2
|
| 58 |
+
1,Female,Low,5.0,Deficient,No,Pneumonia,Klebsiella pneumoniae,RSV,Yes,92.70744076,90.99088026,14,1
|
| 59 |
+
7,Male,High,5.5,Deficient,Yes,Pneumonia,Streptococcus pneumoniae,None,No,99.94270139,98.24850795,2,1
|
| 60 |
+
1,Male,High,44.8,Sufficient,No,Pneumonia,Staphylococcus aureus,RSV,No,61.12605146,59.26551822,9,1
|
| 61 |
+
2,Male,Middle,13.0,Insufficient,Yes,No Pneumonia,Streptococcus pneumoniae,Influenza B,No,83.64980403,77.00659951,2,1
|
| 62 |
+
4,Female,Low,31.6,Sufficient,No,Pneumonia,Staphylococcus aureus,Parainfluenza,No,73.1485804,72.97184557,11,2
|
| 63 |
+
1,Male,Middle,21.2,Insufficient,No,Pneumonia,Streptococcus pneumoniae,None,No,76.9950626,98.8135857,8,2
|
| 64 |
+
11,Female,Low,15.2,Insufficient,No,Severe Pneumonia,Klebsiella pneumoniae,Influenza A,Yes,83.02483836,88.55696632,13,4
|
| 65 |
+
12,Male,High,27.8,Insufficient,Yes,No Pneumonia,Klebsiella pneumoniae,RSV,Yes,73.48736617,58.60794236,3,1
|
| 66 |
+
11,Male,High,5.0,Deficient,Yes,Pneumonia,Staphylococcus aureus,Metapneumovirus,No,96.42236295,96.41900313,2,2
|
| 67 |
+
11,Male,Middle,5.6,Deficient,Yes,No Pneumonia,Streptococcus pneumoniae,Influenza B,Yes,94.59275758,79.75347701,3,1
|
| 68 |
+
9,Male,Low,15.1,Insufficient,Yes,Pneumonia,Klebsiella pneumoniae,Adenovirus,Yes,78.22855992,110.2389734,2,1
|
| 69 |
+
12,Male,High,6.6,Deficient,Yes,Pneumonia,Staphylococcus aureus,RSV,No,92.7927668,111.2853975,4,4
|
| 70 |
+
6,Female,Low,14.2,Insufficient,Yes,Severe Pneumonia,Klebsiella pneumoniae,RSV,Yes,97.19239364,80.47249214,2,5
|
| 71 |
+
13,Female,Low,28.5,Insufficient,No,Pneumonia,Klebsiella pneumoniae,RSV,Yes,57.57997886,53.53466241,8,5
|
| 72 |
+
1,Male,High,21.3,Insufficient,No,No Pneumonia,Klebsiella pneumoniae,RSV,Yes,83.0487022,71.6730305,15,1
|
| 73 |
+
1,Male,Low,33.3,Sufficient,Yes,Severe Pneumonia,Klebsiella pneumoniae,RSV,Yes,71.98562467,65.25207409,2,6
|
| 74 |
+
6,Male,Middle,22.0,Insufficient,No,Pneumonia,None,Influenza A,No,71.2082614,63.90509006,14,1
|
| 75 |
+
4,Male,Middle,29.9,Insufficient,No,No Pneumonia,Staphylococcus aureus,RSV,No,67.75280543,69.54781988,11,1
|
| 76 |
+
4,Female,Middle,19.4,Insufficient,Yes,No Pneumonia,Klebsiella pneumoniae,RSV,Yes,75.59641421,79.09937069,5,1
|
| 77 |
+
11,Male,Middle,14.1,Insufficient,No,Pneumonia,None,Adenovirus,No,95.16982689,86.90123758,13,3
|
| 78 |
+
6,Female,Middle,34.8,Sufficient,No,No Pneumonia,Klebsiella pneumoniae,RSV,No,70.71512633,67.03716142,14,2
|
| 79 |
+
12,Male,Low,12.9,Insufficient,No,Pneumonia,None,Influenza A,No,92.85712687,79.3750592,9,1
|
| 80 |
+
11,Female,Low,23.4,Insufficient,Yes,Severe Pneumonia,Staphylococcus aureus,Influenza A,Yes,77.20323613,69.77268768,3,4
|
| 81 |
+
7,Male,High,27.7,Insufficient,No,Pneumonia,Klebsiella pneumoniae,RSV,No,78.86045975,72.66473732,10,1
|
| 82 |
+
9,Male,Middle,28.4,Insufficient,No,Severe Pneumonia,Staphylococcus aureus,Influenza A,Yes,71.55881716,66.55206296,8,5
|
| 83 |
+
10,Female,Middle,8.3,Deficient,Yes,No Pneumonia,Klebsiella pneumoniae,RSV,No,98.49578814,98.95995815,2,1
|
| 84 |
+
5,Male,Middle,30.5,Sufficient,No,Severe Pneumonia,Klebsiella pneumoniae,RSV,Yes,68.75651788,69.7871564,12,4
|
| 85 |
+
9,Male,Middle,10.2,Insufficient,No,No Pneumonia,Klebsiella pneumoniae,Adenovirus,No,86.40379042,104.7200259,12,1
|
| 86 |
+
4,Male,Low,28.1,Insufficient,No,Severe Pneumonia,Klebsiella pneumoniae,RSV,Yes,80.95475133,78.46385951,11,4
|
| 87 |
+
13,Female,Low,25.4,Insufficient,No,Severe Pneumonia,Klebsiella pneumoniae,Influenza A,No,74.4957562,83.42971096,9,4
|
| 88 |
+
10,Male,Middle,28.2,Insufficient,No,No Pneumonia,Staphylococcus aureus,RSV,No,67.30600046,72.86052854,15,1
|
| 89 |
+
2,Male,Middle,7.9,Deficient,No,Pneumonia,Staphylococcus aureus,RSV,Yes,87.55183051,93.35889201,8,3
|
| 90 |
+
3,Female,Low,25.9,Insufficient,Yes,Severe Pneumonia,Klebsiella pneumoniae,RSV,No,69.56710663,87.95056464,2,4
|
| 91 |
+
13,Female,Middle,30.9,Sufficient,No,No Pneumonia,Staphylococcus aureus,Parainfluenza,Yes,73.32221176,66.07365693,9,2
|
| 92 |
+
11,Female,Low,21.9,Insufficient,No,Pneumonia,Klebsiella pneumoniae,Adenovirus,Yes,77.60568561,82.47423483,8,1
|
| 93 |
+
1,Male,Middle,11.8,Insufficient,No,Pneumonia,Streptococcus pneumoniae,Rhinovirus,No,99.68979488,86.07009974,11,3
|
| 94 |
+
6,Male,High,29.2,Insufficient,No,No Pneumonia,Staphylococcus aureus,Influenza A,Yes,76.73089091,68.65091153,11,1
|
| 95 |
+
13,Male,High,21.1,Insufficient,No,No Pneumonia,Staphylococcus aureus,RSV,No,80.18569065,69.54882987,15,1
|
| 96 |
+
6,Female,Middle,5.0,Deficient,Yes,Severe Pneumonia,Klebsiella pneumoniae,RSV,No,102.0194877,89.05988519,5,6
|
| 97 |
+
1,Male,Middle,11.0,Insufficient,No,Severe Pneumonia,Klebsiella pneumoniae,Influenza A,No,86.52246332,81.57384439,12,4
|
| 98 |
+
11,Male,Low,14.7,Insufficient,No,Pneumonia,Klebsiella pneumoniae,RSV,Yes,76.02607098,90.05495047,9,3
|
| 99 |
+
13,Female,Middle,35.1,Sufficient,No,No Pneumonia,Staphylococcus aureus,Adenovirus,No,59.95208471,73.22164743,13,2
|
| 100 |
+
3,Female,Low,33.3,Sufficient,Yes,Severe Pneumonia,Staphylococcus aureus,RSV,No,67.43603915,65.94197301,5,4
|
| 101 |
+
5,Female,Low,33.3,Sufficient,Yes,Pneumonia,Klebsiella pneumoniae,Influenza B,No,65.58729264,68.53702344,3,1
|
| 102 |
+
3,Female,High,22.3,Insufficient,No,No Pneumonia,Staphylococcus aureus,RSV,No,81.82207694,76.48022916,12,1
|
| 103 |
+
10,Female,Middle,29.1,Insufficient,Yes,No Pneumonia,Klebsiella pneumoniae,Metapneumovirus,No,70.27693109,64.41379264,5,1
|
| 104 |
+
3,Male,High,8.6,Deficient,Yes,No Pneumonia,None,Adenovirus,No,95.14017363,98.2981042,4,1
|
| 105 |
+
10,Female,Middle,20.1,Insufficient,Yes,No Pneumonia,Klebsiella pneumoniae,Adenovirus,No,87.40485815,85.12027755,5,1
|
| 106 |
+
3,Male,Low,24.5,Insufficient,Yes,Severe Pneumonia,Klebsiella pneumoniae,Influenza A,Yes,74.86152075,60.35963212,4,4
|
| 107 |
+
5,Male,High,9.3,Deficient,Yes,No Pneumonia,Klebsiella pneumoniae,RSV,No,93.47572644,88.43512799,3,1
|
| 108 |
+
13,Female,Middle,22.1,Insufficient,No,Severe Pneumonia,Staphylococcus aureus,RSV,Yes,69.1260658,70.84378127,9,6
|
| 109 |
+
5,Female,Low,36.9,Sufficient,Yes,Severe Pneumonia,Klebsiella pneumoniae,RSV,Yes,62.48053011,59.97249144,3,4
|
| 110 |
+
4,Female,Low,25.6,Insufficient,No,Severe Pneumonia,Klebsiella pneumoniae,RSV,No,68.24020937,69.03654746,12,4
|
| 111 |
+
1,Male,Middle,29.2,Insufficient,No,No Pneumonia,Staphylococcus aureus,RSV,Yes,71.52613389,70.42863703,13,2
|
| 112 |
+
5,Female,Low,5.0,Deficient,No,Severe Pneumonia,Klebsiella pneumoniae,RSV,Yes,96.17999878,92.36068212,10,6
|
| 113 |
+
10,Female,Middle,35.6,Sufficient,Yes,No Pneumonia,Klebsiella pneumoniae,RSV,No,65.56338179,61.59087445,3,2
|
| 114 |
+
5,Female,Middle,18.1,Insufficient,No,Pneumonia,Klebsiella pneumoniae,RSV,Yes,79.49938705,90.75910743,10,4
|
| 115 |
+
4,Female,Middle,12.3,Insufficient,No,No Pneumonia,Staphylococcus aureus,RSV,No,99.19234431,92.15742929,15,2
|
| 116 |
+
6,Female,Middle,18.3,Insufficient,No,No Pneumonia,Streptococcus pneumoniae,Influenza B,No,75.35989244,76.0688428,10,1
|
| 117 |
+
10,Male,Middle,5.1,Deficient,Yes,Severe Pneumonia,Klebsiella pneumoniae,RSV,Yes,88.25427086,90.45562614,3,6
|
| 118 |
+
4,Male,Low,30.5,Sufficient,No,Pneumonia,Klebsiella pneumoniae,RSV,No,77.77489188,73.43434444,10,4
|
| 119 |
+
7,Male,High,16.6,Insufficient,Yes,No Pneumonia,None,RSV,No,79.87860435,87.32335657,3,1
|
| 120 |
+
7,Female,Low,12.6,Insufficient,No,Severe Pneumonia,Staphylococcus aureus,Influenza A,No,82.67654656,82.56975809,14,6
|
| 121 |
+
2,Female,Middle,18.3,Insufficient,No,No Pneumonia,Klebsiella pneumoniae,Metapneumovirus,No,93.21455949,79.82807466,8,1
|
| 122 |
+
3,Female,High,31.2,Sufficient,No,No Pneumonia,Staphylococcus aureus,RSV,Yes,73.72861358,83.57444832,14,2
|
| 123 |
+
1,Male,Low,21.3,Insufficient,No,Severe Pneumonia,Klebsiella pneumoniae,RSV,Yes,75.72767726,74.00761529,14,4
|
| 124 |
+
8,Female,Low,5.9,Deficient,Yes,Pneumonia,Klebsiella pneumoniae,Adenovirus,Yes,95.18753953,109.3864367,4,2
|
| 125 |
+
9,Female,Low,24.6,Insufficient,No,Severe Pneumonia,Klebsiella pneumoniae,RSV,Yes,85.12935642,84.22933554,15,6
|
| 126 |
+
4,Female,Low,10.7,Insufficient,Yes,Severe Pneumonia,Klebsiella pneumoniae,Influenza A,Yes,92.7581034,103.1033302,4,6
|
| 127 |
+
6,Male,Low,5.0,Deficient,Yes,Pneumonia,Klebsiella pneumoniae,RSV,Yes,101.4307423,89.96742422,3,2
|
| 128 |
+
9,Female,High,35.0,Sufficient,Yes,No Pneumonia,Klebsiella pneumoniae,Metapneumovirus,No,69.02401589,63.65155301,5,1
|
| 129 |
+
6,Male,Low,24.6,Insufficient,Yes,Severe Pneumonia,Staphylococcus aureus,Influenza A,Yes,80.05126673,71.80668953,2,4
|
| 130 |
+
12,Female,Low,19.2,Insufficient,No,Pneumonia,Staphylococcus aureus,RSV,Yes,80.99870848,89.81402554,14,1
|
| 131 |
+
12,Male,Low,16.5,Insufficient,Yes,Pneumonia,Klebsiella pneumoniae,RSV,Yes,83.55674239,78.95852982,3,3
|
| 132 |
+
12,Male,Low,34.0,Sufficient,Yes,Severe Pneumonia,Klebsiella pneumoniae,RSV,Yes,67.33525193,71.84972395,5,6
|
| 133 |
+
9,Male,Middle,50.9,Sufficient,No,No Pneumonia,Streptococcus pneumoniae,Influenza A,No,52.77226404,54.52639529,10,1
|
| 134 |
+
4,Female,Low,24.5,Insufficient,No,Pneumonia,Staphylococcus aureus,Adenovirus,Yes,74.08342342,75.11576375,13,1
|
| 135 |
+
8,Female,Low,9.7,Deficient,No,Pneumonia,Staphylococcus aureus,RSV,No,90.91026967,100.6892779,15,2
|
| 136 |
+
9,Male,Middle,18.5,Insufficient,No,Severe Pneumonia,Staphylococcus aureus,Influenza A,Yes,79.94827192,82.36143896,15,4
|
| 137 |
+
4,Male,High,24.3,Insufficient,No,Pneumonia,Streptococcus pneumoniae,Metapneumovirus,Yes,76.27842411,82.37268575,11,2
|
| 138 |
+
5,Female,Middle,5.0,Deficient,Yes,Severe Pneumonia,Klebsiella pneumoniae,RSV,Yes,88.74501835,91.04237305,2,5
|
| 139 |
+
8,Female,Middle,23.3,Insufficient,Yes,Severe Pneumonia,Klebsiella pneumoniae,RSV,Yes,89.72988733,70.91250549,3,6
|
| 140 |
+
12,Male,Middle,24.4,Insufficient,No,No Pneumonia,Streptococcus pneumoniae,RSV,No,71.93529941,81.44916423,15,2
|
| 141 |
+
10,Female,Middle,33.3,Sufficient,Yes,Severe Pneumonia,Staphylococcus aureus,Influenza A,Yes,64.89269638,70.14672195,3,5
|
| 142 |
+
6,Female,Middle,32.2,Sufficient,No,Pneumonia,Staphylococcus aureus,RSV,Yes,63.70367803,69.74646131,11,5
|
Output1.spv
ADDED
|
@@ -0,0 +1,3 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
version https://git-lfs.github.com/spec/v1
|
| 2 |
+
oid sha256:d9da88cc8ebc852410f732aa6af729a72158f5fc35c393b053e57ef7f3618a91
|
| 3 |
+
size 190912
|
Untitled9.sav
ADDED
|
Binary file (21.6 kB). View file
|
|
|
actual_vs_predicted_heatmap.png
ADDED

|
app.py
ADDED
|
@@ -0,0 +1,65 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
import gradio as gr
|
| 2 |
+
import pandas as pd
|
| 3 |
+
import numpy as np
|
| 4 |
+
import joblib
|
| 5 |
+
|
| 6 |
+
# Load trained components
|
| 7 |
+
model = joblib.load("voting_model_multiclass.pkl")
|
| 8 |
+
scaler = joblib.load("scaler.pkl")
|
| 9 |
+
label_encoders = joblib.load("feature_label_encoders.pkl")
|
| 10 |
+
target_encoder = joblib.load("target_label_encoder.pkl")
|
| 11 |
+
|
| 12 |
+
# Feature order used during training
|
| 13 |
+
feature_order = [
|
| 14 |
+
'Age', 'Sex', 'Socioeconomic_Status', 'Vitamin_D_Level_ng/ml',
|
| 15 |
+
'Vitamin_D_Status', 'Vitamin_D_Supplemented', 'Bacterial_Infection',
|
| 16 |
+
'Viral_Infection', 'Co_Infection', 'IL6_pg/ml', 'IL8_pg/ml'
|
| 17 |
+
]
|
| 18 |
+
|
| 19 |
+
def predict_arti_severity(
|
| 20 |
+
age, sex, ses, vit_d_level, vit_d_status, vit_d_supp,
|
| 21 |
+
bacterial, viral, co_infect, il6, il8
|
| 22 |
+
):
|
| 23 |
+
# Create a single row DataFrame
|
| 24 |
+
input_data = pd.DataFrame([[
|
| 25 |
+
age, sex, ses, vit_d_level, vit_d_status, vit_d_supp,
|
| 26 |
+
bacterial, viral, co_infect, il6, il8
|
| 27 |
+
]], columns=feature_order)
|
| 28 |
+
|
| 29 |
+
# Encode categorical columns
|
| 30 |
+
for col in input_data.select_dtypes(include='object').columns:
|
| 31 |
+
le = label_encoders[col]
|
| 32 |
+
input_data[col] = le.transform(input_data[col])
|
| 33 |
+
|
| 34 |
+
# Scale the input
|
| 35 |
+
input_scaled = scaler.transform(input_data)
|
| 36 |
+
|
| 37 |
+
# Predict
|
| 38 |
+
pred = model.predict(input_scaled)[0]
|
| 39 |
+
pred_label = target_encoder.inverse_transform([pred])[0]
|
| 40 |
+
|
| 41 |
+
return f"Predicted ARTI Severity: {pred_label}"
|
| 42 |
+
|
| 43 |
+
# Define Gradio interface
|
| 44 |
+
interface = gr.Interface(
|
| 45 |
+
fn=predict_arti_severity,
|
| 46 |
+
inputs=[
|
| 47 |
+
gr.Number(label="Age (years)", value=2),
|
| 48 |
+
gr.Radio(choices=['Male', 'Female'], label="Sex"),
|
| 49 |
+
gr.Radio(choices=['Low', 'Middle', 'High'], label="Socioeconomic Status"),
|
| 50 |
+
gr.Number(label="Vitamin D Level (ng/ml)", value=20),
|
| 51 |
+
gr.Radio(choices=['Deficient', 'Insufficient', 'Sufficient'], label="Vitamin D Status"),
|
| 52 |
+
gr.Radio(choices=['Yes', 'No'], label="Vitamin D Supplemented"),
|
| 53 |
+
gr.Dropdown(choices=['Streptococcus pneumoniae', 'Klebsiella pneumoniae', 'Staphylococcus aureus', 'None'], label="Bacterial Infection"),
|
| 54 |
+
gr.Dropdown(choices=['RSV', 'Influenza A', 'Influenza B', 'Adenovirus', 'Rhinovirus', 'Metapneumovirus', 'Parainfluenza', 'None'], label="Viral Infection"),
|
| 55 |
+
gr.Radio(choices=['Yes', 'No'], label="Co-Infection"),
|
| 56 |
+
gr.Number(label="IL-6 (pg/ml)", value=25),
|
| 57 |
+
gr.Number(label="IL-8 (pg/ml)", value=40)
|
| 58 |
+
],
|
| 59 |
+
outputs=gr.Textbox(label="Prediction Result"),
|
| 60 |
+
title="ARTI Severity Predictor",
|
| 61 |
+
description="Predict ARTI severity in children based on vitamin D, infection data, and inflammation markers."
|
| 62 |
+
)
|
| 63 |
+
|
| 64 |
+
# Launch app
|
| 65 |
+
interface.launch()
|
class_distribution.png
ADDED
|
confusion_matrix.png
ADDED

|
confusion_matrix_multiclass.png
ADDED

|
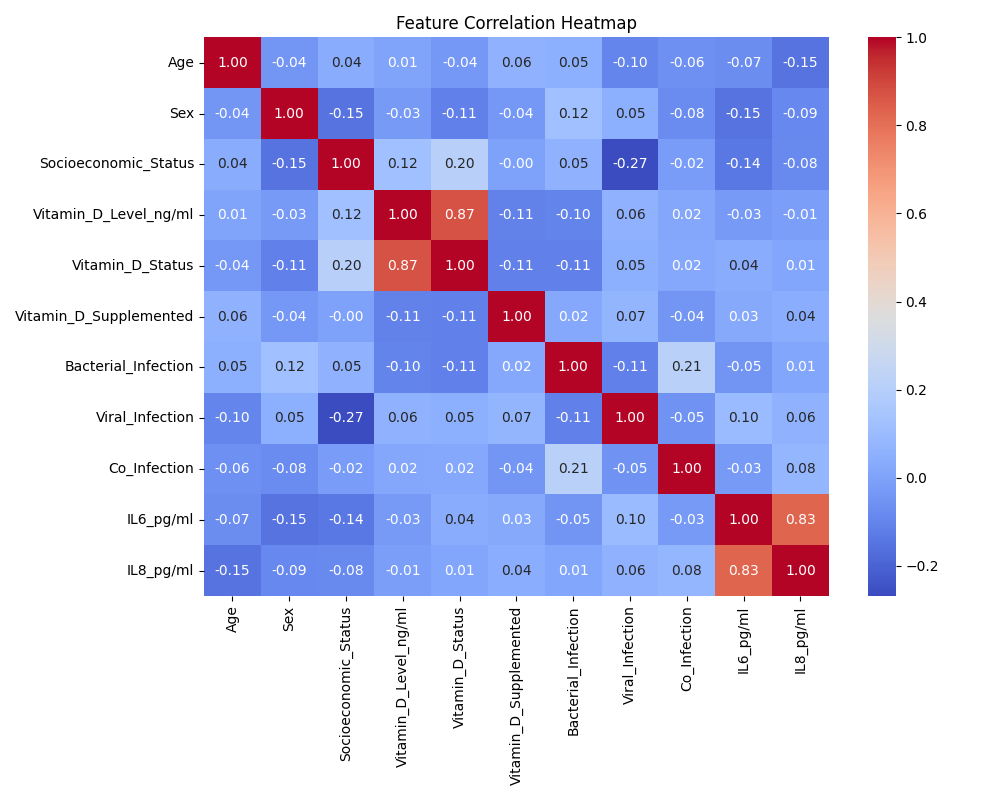
correlation_heatmap.png
ADDED
|
Git LFS Details
|
feature_importance.png
ADDED
|
feature_importance_rf.png
ADDED
|
feature_label_encoders.pkl
ADDED
|
@@ -0,0 +1,3 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
version https://git-lfs.github.com/spec/v1
|
| 2 |
+
oid sha256:a20d567cf177ba82f39df49bc609b09d511e41942f93de251fdb1b2873008e62
|
| 3 |
+
size 2553
|
main.py
ADDED
|
@@ -0,0 +1,146 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
import pandas as pd
|
| 2 |
+
import numpy as np
|
| 3 |
+
import matplotlib.pyplot as plt
|
| 4 |
+
import seaborn as sns
|
| 5 |
+
import joblib
|
| 6 |
+
from sklearn.model_selection import train_test_split
|
| 7 |
+
from sklearn.preprocessing import LabelEncoder, StandardScaler
|
| 8 |
+
from sklearn.ensemble import RandomForestClassifier, VotingClassifier
|
| 9 |
+
from sklearn.linear_model import LogisticRegression
|
| 10 |
+
from sklearn.svm import SVC
|
| 11 |
+
from sklearn.metrics import (
|
| 12 |
+
classification_report, confusion_matrix, accuracy_score,
|
| 13 |
+
ConfusionMatrixDisplay
|
| 14 |
+
)
|
| 15 |
+
import warnings
|
| 16 |
+
warnings.filterwarnings('ignore')
|
| 17 |
+
|
| 18 |
+
# =====================
|
| 19 |
+
# 1. Load Dataset
|
| 20 |
+
# =====================
|
| 21 |
+
df = pd.read_csv("ARTI_Main_Data.csv")
|
| 22 |
+
|
| 23 |
+
# Handle missing values
|
| 24 |
+
df['Bacterial_Infection'] = df['Bacterial_Infection'].fillna("None")
|
| 25 |
+
df['Viral_Infection'] = df['Viral_Infection'].fillna("None")
|
| 26 |
+
|
| 27 |
+
# =====================
|
| 28 |
+
# 2. Set up features and multi-class target
|
| 29 |
+
# =====================
|
| 30 |
+
features = [
|
| 31 |
+
'Age', 'Sex', 'Socioeconomic_Status', 'Vitamin_D_Level_ng/ml',
|
| 32 |
+
'Vitamin_D_Status', 'Vitamin_D_Supplemented', 'Bacterial_Infection',
|
| 33 |
+
'Viral_Infection', 'Co_Infection', 'IL6_pg/ml', 'IL8_pg/ml'
|
| 34 |
+
]
|
| 35 |
+
target = 'ARTI_Severity'
|
| 36 |
+
|
| 37 |
+
# =====================
|
| 38 |
+
# 3. Encode features and target
|
| 39 |
+
# =====================
|
| 40 |
+
df_encoded = df[features].copy()
|
| 41 |
+
cat_cols = df_encoded.select_dtypes(include=['object']).columns
|
| 42 |
+
label_encoders = {}
|
| 43 |
+
|
| 44 |
+
for col in cat_cols:
|
| 45 |
+
le = LabelEncoder()
|
| 46 |
+
df_encoded[col] = le.fit_transform(df_encoded[col])
|
| 47 |
+
label_encoders[col] = le
|
| 48 |
+
|
| 49 |
+
# Encode target (multi-class)
|
| 50 |
+
target_encoder = LabelEncoder()
|
| 51 |
+
df['ARTI_Severity_Label'] = target_encoder.fit_transform(df[target])
|
| 52 |
+
y = df['ARTI_Severity_Label']
|
| 53 |
+
|
| 54 |
+
# =====================
|
| 55 |
+
# 4. Scale numerical features
|
| 56 |
+
# =====================
|
| 57 |
+
scaler = StandardScaler()
|
| 58 |
+
X_scaled = scaler.fit_transform(df_encoded)
|
| 59 |
+
|
| 60 |
+
# =====================
|
| 61 |
+
# 5. Train-test split
|
| 62 |
+
# =====================
|
| 63 |
+
X_train, X_test, y_train, y_test = train_test_split(
|
| 64 |
+
X_scaled, y, test_size=0.2, random_state=42, stratify=y
|
| 65 |
+
)
|
| 66 |
+
|
| 67 |
+
# =====================
|
| 68 |
+
# 6. Define Models
|
| 69 |
+
# =====================
|
| 70 |
+
log_reg = LogisticRegression(max_iter=500)
|
| 71 |
+
rf = RandomForestClassifier(n_estimators=100, random_state=42)
|
| 72 |
+
svm = SVC(probability=True)
|
| 73 |
+
|
| 74 |
+
# Voting classifier
|
| 75 |
+
voting_model = VotingClassifier(estimators=[
|
| 76 |
+
('lr', log_reg),
|
| 77 |
+
('rf', rf),
|
| 78 |
+
('svm', svm)
|
| 79 |
+
], voting='hard')
|
| 80 |
+
|
| 81 |
+
# =====================
|
| 82 |
+
# 7. Train Model
|
| 83 |
+
# =====================
|
| 84 |
+
voting_model.fit(X_train, y_train)
|
| 85 |
+
|
| 86 |
+
# Save model and preprocessors
|
| 87 |
+
joblib.dump(voting_model, "voting_model_multiclass.pkl")
|
| 88 |
+
joblib.dump(scaler, "scaler.pkl")
|
| 89 |
+
joblib.dump(label_encoders, "feature_label_encoders.pkl")
|
| 90 |
+
joblib.dump(target_encoder, "target_label_encoder.pkl")
|
| 91 |
+
|
| 92 |
+
# =====================
|
| 93 |
+
# 8. Evaluation
|
| 94 |
+
# =====================
|
| 95 |
+
y_pred = voting_model.predict(X_test)
|
| 96 |
+
|
| 97 |
+
print("\n📊 Confusion Matrix:\n", confusion_matrix(y_test, y_pred))
|
| 98 |
+
print("\n📑 Classification Report:\n", classification_report(y_test, y_pred, target_names=target_encoder.classes_))
|
| 99 |
+
print("\n✅ Accuracy Score:", accuracy_score(y_test, y_pred))
|
| 100 |
+
|
| 101 |
+
# =====================
|
| 102 |
+
# 9. Visualizations
|
| 103 |
+
# =====================
|
| 104 |
+
|
| 105 |
+
# 1. Confusion Matrix
|
| 106 |
+
disp = ConfusionMatrixDisplay(confusion_matrix=confusion_matrix(y_test, y_pred),
|
| 107 |
+
display_labels=target_encoder.classes_)
|
| 108 |
+
disp.plot(cmap=plt.cm.Blues)
|
| 109 |
+
plt.title("Confusion Matrix")
|
| 110 |
+
plt.savefig("confusion_matrix_multiclass.png")
|
| 111 |
+
plt.show()
|
| 112 |
+
|
| 113 |
+
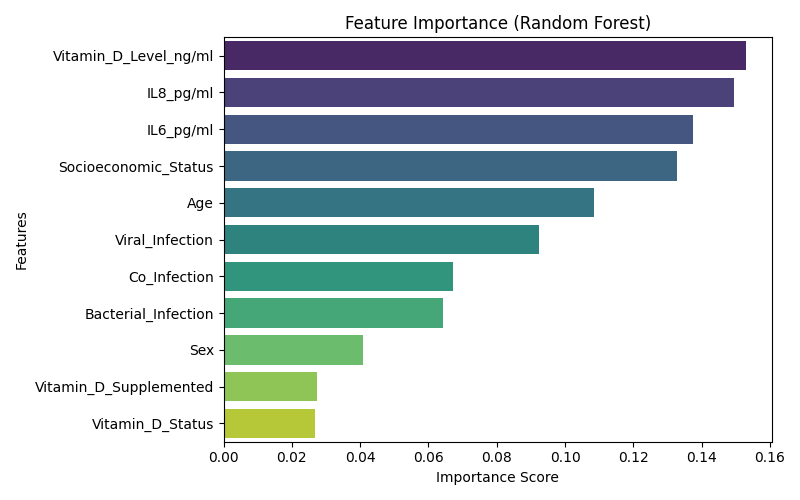
# 2. Feature Importance (fit rf separately for this)
|
| 114 |
+
rf.fit(X_train, y_train)
|
| 115 |
+
plt.figure(figsize=(8, 5))
|
| 116 |
+
importances = rf.feature_importances_
|
| 117 |
+
indices = np.argsort(importances)[::-1]
|
| 118 |
+
feature_names = df_encoded.columns
|
| 119 |
+
|
| 120 |
+
sns.barplot(x=importances[indices], y=np.array(feature_names)[indices], palette='viridis')
|
| 121 |
+
plt.title("Feature Importance (Random Forest)")
|
| 122 |
+
plt.xlabel("Importance Score")
|
| 123 |
+
plt.ylabel("Features")
|
| 124 |
+
plt.tight_layout()
|
| 125 |
+
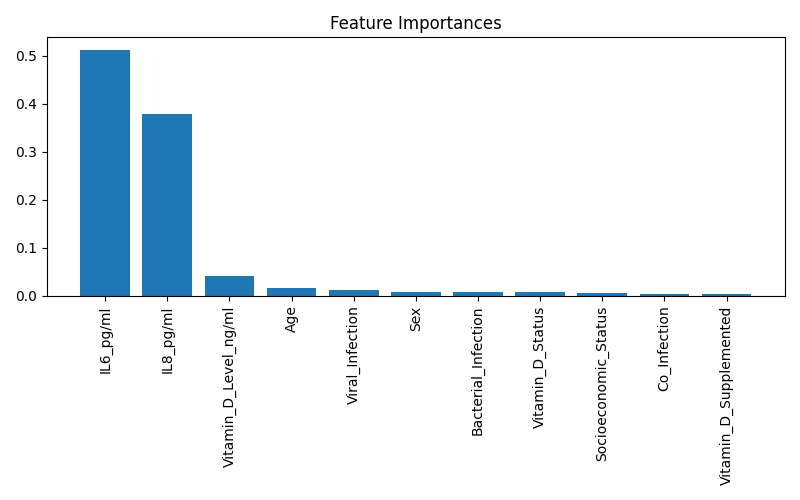
plt.savefig("feature_importance_rf.png")
|
| 126 |
+
plt.show()
|
| 127 |
+
|
| 128 |
+
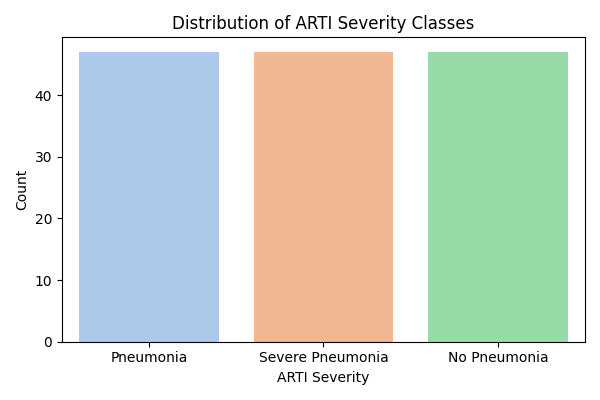
# 3. Class Distribution
|
| 129 |
+
plt.figure(figsize=(6, 4))
|
| 130 |
+
sns.countplot(x=df[target], palette='pastel')
|
| 131 |
+
plt.title("Distribution of ARTI Severity Classes")
|
| 132 |
+
plt.xlabel("ARTI Severity")
|
| 133 |
+
plt.ylabel("Count")
|
| 134 |
+
plt.tight_layout()
|
| 135 |
+
plt.savefig("class_distribution.png")
|
| 136 |
+
plt.show()
|
| 137 |
+
|
| 138 |
+
# 4. Actual vs Predicted Comparison
|
| 139 |
+
plt.figure(figsize=(8, 5))
|
| 140 |
+
sns.heatmap(confusion_matrix(y_test, y_pred), annot=True, fmt="d", cmap="YlGnBu",
|
| 141 |
+
xticklabels=target_encoder.classes_, yticklabels=target_encoder.classes_)
|
| 142 |
+
plt.xlabel("Predicted")
|
| 143 |
+
plt.ylabel("Actual")
|
| 144 |
+
plt.title("Actual vs Predicted Heatmap")
|
| 145 |
+
plt.savefig("actual_vs_predicted_heatmap.png")
|
| 146 |
+
plt.show()
|
mh.py
ADDED
|
@@ -0,0 +1,65 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
import gradio as gr
|
| 2 |
+
import pandas as pd
|
| 3 |
+
import numpy as np
|
| 4 |
+
import joblib
|
| 5 |
+
|
| 6 |
+
# Load trained components
|
| 7 |
+
model = joblib.load("voting_model_multiclass.pkl")
|
| 8 |
+
scaler = joblib.load("scaler.pkl")
|
| 9 |
+
label_encoders = joblib.load("feature_label_encoders.pkl")
|
| 10 |
+
target_encoder = joblib.load("target_label_encoder.pkl")
|
| 11 |
+
|
| 12 |
+
# Feature order used during training
|
| 13 |
+
feature_order = [
|
| 14 |
+
'Age', 'Sex', 'Socioeconomic_Status', 'Vitamin_D_Level_ng/ml',
|
| 15 |
+
'Vitamin_D_Status', 'Vitamin_D_Supplemented', 'Bacterial_Infection',
|
| 16 |
+
'Viral_Infection', 'Co_Infection', 'IL6_pg/ml', 'IL8_pg/ml'
|
| 17 |
+
]
|
| 18 |
+
|
| 19 |
+
def predict_arti_severity(
|
| 20 |
+
age, sex, ses, vit_d_level, vit_d_status, vit_d_supp,
|
| 21 |
+
bacterial, viral, co_infect, il6, il8
|
| 22 |
+
):
|
| 23 |
+
# Create a single row DataFrame
|
| 24 |
+
input_data = pd.DataFrame([[
|
| 25 |
+
age, sex, ses, vit_d_level, vit_d_status, vit_d_supp,
|
| 26 |
+
bacterial, viral, co_infect, il6, il8
|
| 27 |
+
]], columns=feature_order)
|
| 28 |
+
|
| 29 |
+
# Encode categorical columns
|
| 30 |
+
for col in input_data.select_dtypes(include='object').columns:
|
| 31 |
+
le = label_encoders[col]
|
| 32 |
+
input_data[col] = le.transform(input_data[col])
|
| 33 |
+
|
| 34 |
+
# Scale the input
|
| 35 |
+
input_scaled = scaler.transform(input_data)
|
| 36 |
+
|
| 37 |
+
# Predict
|
| 38 |
+
pred = model.predict(input_scaled)[0]
|
| 39 |
+
pred_label = target_encoder.inverse_transform([pred])[0]
|
| 40 |
+
|
| 41 |
+
return f"Predicted ARTI Severity: {pred_label}"
|
| 42 |
+
|
| 43 |
+
# Define Gradio interface
|
| 44 |
+
interface = gr.Interface(
|
| 45 |
+
fn=predict_arti_severity,
|
| 46 |
+
inputs=[
|
| 47 |
+
gr.Number(label="Age (years)", value=2),
|
| 48 |
+
gr.Radio(choices=['Male', 'Female'], label="Sex"),
|
| 49 |
+
gr.Radio(choices=['Low', 'Middle', 'High'], label="Socioeconomic Status"),
|
| 50 |
+
gr.Number(label="Vitamin D Level (ng/ml)", value=20),
|
| 51 |
+
gr.Radio(choices=['Deficient', 'Insufficient', 'Sufficient'], label="Vitamin D Status"),
|
| 52 |
+
gr.Radio(choices=['Yes', 'No'], label="Vitamin D Supplemented"),
|
| 53 |
+
gr.Dropdown(choices=['Streptococcus pneumoniae', 'Klebsiella pneumoniae', 'Staphylococcus aureus', 'None'], label="Bacterial Infection"),
|
| 54 |
+
gr.Dropdown(choices=['RSV', 'Influenza A', 'Influenza B', 'Adenovirus', 'Rhinovirus', 'Metapneumovirus', 'Parainfluenza', 'None'], label="Viral Infection"),
|
| 55 |
+
gr.Radio(choices=['Yes', 'No'], label="Co-Infection"),
|
| 56 |
+
gr.Number(label="IL-6 (pg/ml)", value=25),
|
| 57 |
+
gr.Number(label="IL-8 (pg/ml)", value=40)
|
| 58 |
+
],
|
| 59 |
+
outputs=gr.Textbox(label="Prediction Result"),
|
| 60 |
+
title="ARTI Severity Predictor",
|
| 61 |
+
description="Predict ARTI severity in children based on vitamin D, infection data, and inflammation markers."
|
| 62 |
+
)
|
| 63 |
+
|
| 64 |
+
# Launch app
|
| 65 |
+
interface.launch()
|
roc_curve.png
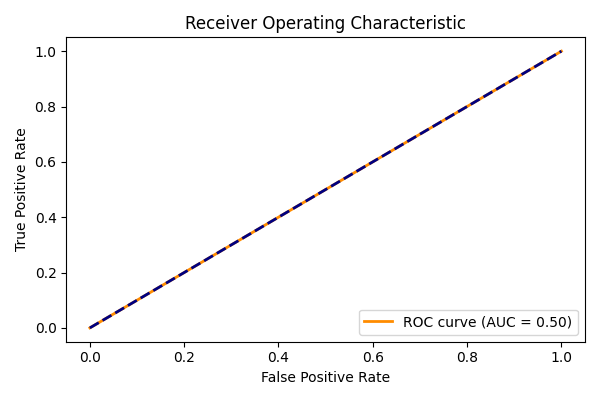
ADDED
|
scaler.pkl
ADDED
|
@@ -0,0 +1,3 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
version https://git-lfs.github.com/spec/v1
|
| 2 |
+
oid sha256:e536ef2a51a198f4421e113252d7aca4a003bc63a7c14e69a79dbb2185a27bdc
|
| 3 |
+
size 1391
|
target_label_encoder.pkl
ADDED
|
@@ -0,0 +1,3 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
version https://git-lfs.github.com/spec/v1
|
| 2 |
+
oid sha256:81434ab47403fba3acdbb129593a5efad043bd7f18c3dde8886d639d3cfa4b5f
|
| 3 |
+
size 576
|
voting_model.pkl
ADDED
|
@@ -0,0 +1,3 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
version https://git-lfs.github.com/spec/v1
|
| 2 |
+
oid sha256:c923864f3379b66f44b2ef026cc7a2dbf8d945084f53bda13dfb64f122f8821a
|
| 3 |
+
size 148520
|
voting_model_multiclass.pkl
ADDED
|
@@ -0,0 +1,3 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
version https://git-lfs.github.com/spec/v1
|
| 2 |
+
oid sha256:f39569fb33021aa87a588940ff07beef398a48b2144e1564997620adca07e14e
|
| 3 |
+
size 699792
|