Spaces:
Runtime error
Runtime error
Tsumugii24
commited on
Commit
·
dec70f4
1
Parent(s):
a99dcc4
add project files
Browse filesThis view is limited to 50 files because it contains too many changes.
See raw diff
- .gitignore +54 -0
- LICENSE +21 -0
- README.md +222 -13
- app.py +546 -0
- cls_name/cls_name_cells_en.yaml +1 -0
- cls_name/cls_name_cells_zh.yaml +1 -0
- favicons/Lesion-Cells DET.png +0 -0
- favicons/logo.ico +0 -0
- img_examples/ground_truth_label/carcinoma_in_situ/149315671-149315740-001.BMP +0 -0
- img_examples/ground_truth_label/carcinoma_in_situ/149315671-149315740-002.BMP +0 -0
- img_examples/ground_truth_label/carcinoma_in_situ/149315671-149315740-003.BMP +0 -0
- img_examples/ground_truth_label/carcinoma_in_situ/149315671-149315740-004.BMP +0 -0
- img_examples/ground_truth_label/carcinoma_in_situ/149315671-149315749-001.BMP +0 -0
- img_examples/ground_truth_label/carcinoma_in_situ/149315671-149315749-002.BMP +0 -0
- img_examples/ground_truth_label/carcinoma_in_situ/149315775-149315790-001.BMP +0 -0
- img_examples/ground_truth_label/carcinoma_in_situ/149315775-149315790-002.BMP +0 -0
- img_examples/ground_truth_label/carcinoma_in_situ/149315775-149315790-003.BMP +0 -0
- img_examples/ground_truth_label/carcinoma_in_situ/149316025-149316032-001.BMP +0 -0
- img_examples/ground_truth_label/carcinoma_in_situ/149316117-149316122-002.BMP +0 -0
- img_examples/ground_truth_label/carcinoma_in_situ/149316117-149316122-003.BMP +0 -0
- img_examples/ground_truth_label/carcinoma_in_situ/149316117-149316122-004.BMP +0 -0
- img_examples/ground_truth_label/carcinoma_in_situ/149316117-149316131-001.BMP +0 -0
- img_examples/ground_truth_label/carcinoma_in_situ/149316117-149316131-002.BMP +0 -0
- img_examples/ground_truth_label/carcinoma_in_situ/149316426-149316462-001.BMP +0 -0
- img_examples/ground_truth_label/carcinoma_in_situ/149316426-149316462-002.BMP +0 -0
- img_examples/ground_truth_label/carcinoma_in_situ/149316426-149316462-003.BMP +0 -0
- img_examples/ground_truth_label/carcinoma_in_situ/149316426-149316462-004.BMP +0 -0
- img_examples/ground_truth_label/carcinoma_in_situ/149317114-149317152-001.BMP +0 -0
- img_examples/ground_truth_label/carcinoma_in_situ/149317114-149317152-002.BMP +0 -0
- img_examples/ground_truth_label/carcinoma_in_situ/149317114-149317152-004.BMP +0 -0
- img_examples/ground_truth_label/carcinoma_in_situ/149357956-149358043-001.BMP +0 -0
- img_examples/ground_truth_label/carcinoma_in_situ/153826597-153826619-001.BMP +0 -0
- img_examples/ground_truth_label/carcinoma_in_situ/153826597-153826619-002.BMP +0 -0
- img_examples/ground_truth_label/carcinoma_in_situ/153827595-153827657-001.BMP +0 -0
- img_examples/ground_truth_label/carcinoma_in_situ/153827595-153827657-002.BMP +0 -0
- img_examples/ground_truth_label/carcinoma_in_situ/153827595-153827664-001.BMP +0 -0
- img_examples/ground_truth_label/carcinoma_in_situ/153827595-153827669-001.BMP +0 -0
- img_examples/ground_truth_label/carcinoma_in_situ/153830680-153830693-001.BMP +0 -0
- img_examples/ground_truth_label/carcinoma_in_situ/153831027-153831036-001.BMP +0 -0
- img_examples/ground_truth_label/carcinoma_in_situ/153831027-153831036-002.BMP +0 -0
- img_examples/ground_truth_label/carcinoma_in_situ/153831027-153831045-001.BMP +0 -0
- img_examples/ground_truth_label/carcinoma_in_situ/153831027-153831045-002.BMP +0 -0
- img_examples/ground_truth_label/carcinoma_in_situ/153831027-153831045-003.BMP +0 -0
- img_examples/ground_truth_label/carcinoma_in_situ/153831426-153831438-001.BMP +0 -0
- img_examples/ground_truth_label/carcinoma_in_situ/153831471-153831479-001.BMP +0 -0
- img_examples/ground_truth_label/carcinoma_in_situ/153831471-153831486-001.BMP +0 -0
- img_examples/ground_truth_label/carcinoma_in_situ/153831471-153831486-002.BMP +0 -0
- img_examples/ground_truth_label/carcinoma_in_situ/153831471-153831486-003.BMP +0 -0
- img_examples/ground_truth_label/carcinoma_in_situ/153831471-153831486-004.BMP +0 -0
- img_examples/ground_truth_label/carcinoma_in_situ/153916114-153916150-001.BMP +0 -0
.gitignore
ADDED
|
@@ -0,0 +1,54 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
# video
|
| 2 |
+
*.mp4
|
| 3 |
+
*.avi
|
| 4 |
+
.ipynb_checkpoints
|
| 5 |
+
/__pycache__
|
| 6 |
+
*/__pycache__
|
| 7 |
+
|
| 8 |
+
# log
|
| 9 |
+
*.log
|
| 10 |
+
*.data
|
| 11 |
+
*.txt
|
| 12 |
+
*.csv
|
| 13 |
+
|
| 14 |
+
|
| 15 |
+
# hyper
|
| 16 |
+
# *.yaml
|
| 17 |
+
*.json
|
| 18 |
+
|
| 19 |
+
# compressed
|
| 20 |
+
*.zip
|
| 21 |
+
*.tar
|
| 22 |
+
*.tar.gz
|
| 23 |
+
*.rar
|
| 24 |
+
|
| 25 |
+
# font
|
| 26 |
+
/fonts
|
| 27 |
+
*.ttc
|
| 28 |
+
*.ttf
|
| 29 |
+
*.otf
|
| 30 |
+
*.pkl
|
| 31 |
+
|
| 32 |
+
# model
|
| 33 |
+
*.db
|
| 34 |
+
*.pt
|
| 35 |
+
|
| 36 |
+
# ide
|
| 37 |
+
.idea/
|
| 38 |
+
.vscode/
|
| 39 |
+
|
| 40 |
+
|
| 41 |
+
/flagged
|
| 42 |
+
/run
|
| 43 |
+
!requirements.txt
|
| 44 |
+
!cls_name/*
|
| 45 |
+
!model_config/*
|
| 46 |
+
!img_examples/*
|
| 47 |
+
!video_examples/*
|
| 48 |
+
!README.md
|
| 49 |
+
|
| 50 |
+
# testing
|
| 51 |
+
!app.py
|
| 52 |
+
test.py
|
| 53 |
+
test*.py
|
| 54 |
+
|
LICENSE
ADDED
|
@@ -0,0 +1,21 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
MIT License
|
| 2 |
+
|
| 3 |
+
Copyright (c) 2023 Tsumugii
|
| 4 |
+
|
| 5 |
+
Permission is hereby granted, free of charge, to any person obtaining a copy
|
| 6 |
+
of this software and associated documentation files (the "Software"), to deal
|
| 7 |
+
in the Software without restriction, including without limitation the rights
|
| 8 |
+
to use, copy, modify, merge, publish, distribute, sublicense, and/or sell
|
| 9 |
+
copies of the Software, and to permit persons to whom the Software is
|
| 10 |
+
furnished to do so, subject to the following conditions:
|
| 11 |
+
|
| 12 |
+
The above copyright notice and this permission notice shall be included in all
|
| 13 |
+
copies or substantial portions of the Software.
|
| 14 |
+
|
| 15 |
+
THE SOFTWARE IS PROVIDED "AS IS", WITHOUT WARRANTY OF ANY KIND, EXPRESS OR
|
| 16 |
+
IMPLIED, INCLUDING BUT NOT LIMITED TO THE WARRANTIES OF MERCHANTABILITY,
|
| 17 |
+
FITNESS FOR A PARTICULAR PURPOSE AND NONINFRINGEMENT. IN NO EVENT SHALL THE
|
| 18 |
+
AUTHORS OR COPYRIGHT HOLDERS BE LIABLE FOR ANY CLAIM, DAMAGES OR OTHER
|
| 19 |
+
LIABILITY, WHETHER IN AN ACTION OF CONTRACT, TORT OR OTHERWISE, ARISING FROM,
|
| 20 |
+
OUT OF OR IN CONNECTION WITH THE SOFTWARE OR THE USE OR OTHER DEALINGS IN THE
|
| 21 |
+
SOFTWARE.
|
README.md
CHANGED
|
@@ -1,13 +1,222 @@
|
|
| 1 |
-
|
| 2 |
-
|
| 3 |
-
|
| 4 |
-
|
| 5 |
-
|
| 6 |
-
|
| 7 |
-
|
| 8 |
-
|
| 9 |
-
|
| 10 |
-
|
| 11 |
-
|
| 12 |
-
|
| 13 |
-
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+

|
| 2 |
+
|
| 3 |
+
</div>
|
| 4 |
+
|
| 5 |
+
### <div align="center"><h2>Description</h2></div>
|
| 6 |
+
|
| 7 |
+
**Lesion-Cells DET** stands for Multi-granularity **Lesion Cells Detection**.
|
| 8 |
+
|
| 9 |
+
The projects employs both CNN-based and Transformer-based neural networks for Object Detection.
|
| 10 |
+
|
| 11 |
+
The system excels at detecting 7 types of cells with varying granularity in images. Additionally, it provides statistical information on the relative sizes and lesion degree distribution ratios of the identified cells.
|
| 12 |
+
|
| 13 |
+
|
| 14 |
+
|
| 15 |
+
</div>
|
| 16 |
+
|
| 17 |
+
### <div align="center"><h2>Acknowledgements</h2></div>
|
| 18 |
+
|
| 19 |
+
***I would like to express my sincere gratitude to Professor Lio for his invaluable guidance in Office Hour and supports throughout the development of this project. Professor's expertise and insightful feedback played a crucial role in shaping the direction of the project.***
|
| 20 |
+
|
| 21 |
+

|
| 22 |
+
|
| 23 |
+
</div>
|
| 24 |
+
|
| 25 |
+
### <div align="center"><h2>Demonstration</h2></div>
|
| 26 |
+
|
| 27 |
+

|
| 28 |
+
|
| 29 |
+
|
| 30 |
+
|
| 31 |
+
|
| 32 |
+
|
| 33 |
+
</div>
|
| 34 |
+
|
| 35 |
+
### <div align="center"><h2>ToDo</h2></div>
|
| 36 |
+
|
| 37 |
+
- [x] ~~Change the large weights files with Google Drive sharing link~~
|
| 38 |
+
- [x] ~~Add Professor Lio's brief introduction~~
|
| 39 |
+
- [x] ~~Add a .gif demonstration instead of a static image~~
|
| 40 |
+
- [ ] deploy the demo on HuggingFace
|
| 41 |
+
- [ ] Train models that have better performance
|
| 42 |
+
- [ ] Upload part of the datasets, so that everyone can train their own customized models
|
| 43 |
+
|
| 44 |
+
|
| 45 |
+
|
| 46 |
+
|
| 47 |
+
|
| 48 |
+
</div>
|
| 49 |
+
|
| 50 |
+
### <div align="center"><h2>Quick Start</h2></div>
|
| 51 |
+
|
| 52 |
+
<details open>
|
| 53 |
+
<summary><h4>Installation</h4></summary>
|
| 54 |
+
|
| 55 |
+
*I strongly recommend you to use **conda**. Both Anaconda and miniconda is OK!*
|
| 56 |
+
|
| 57 |
+
|
| 58 |
+
1. create a virtual **conda** environment for the demo 😆
|
| 59 |
+
|
| 60 |
+
```bash
|
| 61 |
+
$ conda create -n demo python==3.8
|
| 62 |
+
$ conda activate demo
|
| 63 |
+
```
|
| 64 |
+
|
| 65 |
+
2. install essential **requirements** by run the following command in the CLI 😊
|
| 66 |
+
|
| 67 |
+
```bash
|
| 68 |
+
$ git clone https://github.com/Tsumugii24/lesion-cells-det
|
| 69 |
+
$ cd lesion-cells-det
|
| 70 |
+
$ pip install -r requirements.txt
|
| 71 |
+
```
|
| 72 |
+
|
| 73 |
+
3. download the **weights** files from Google Drive that have already been trained properly
|
| 74 |
+
|
| 75 |
+
here is the link, from where you can download your preferred model and then test its performance 🤗
|
| 76 |
+
|
| 77 |
+
```html
|
| 78 |
+
https://drive.google.com/drive/folders/1-H4nN8viLdH6nniuiGO-_wJDENDf-BkL?usp=sharing
|
| 79 |
+
```
|
| 80 |
+
|
| 81 |
+
|
| 82 |
+
|
| 83 |
+
4. remember to put the weights files under the root of the project 😉
|
| 84 |
+
|
| 85 |
+
</details>
|
| 86 |
+
|
| 87 |
+
<details open>
|
| 88 |
+
<summary><h4>Run</h4></summary>
|
| 89 |
+
|
| 90 |
+
```bash
|
| 91 |
+
$ python gradio_demo.py
|
| 92 |
+
```
|
| 93 |
+
|
| 94 |
+
Now, if everything is OK, your default browser will open automatically, and Gradio is running on local URL: http://127.0.0.1:7860
|
| 95 |
+
|
| 96 |
+
</details>
|
| 97 |
+
|
| 98 |
+
<details open>
|
| 99 |
+
<summary><h4>Datasets</h4></summary>
|
| 100 |
+
|
| 101 |
+
The original datasets origins from **Kaggle**, **iFLYTEK AI algorithm competition** and **other open source** sources.
|
| 102 |
+
|
| 103 |
+
Anyway, we annotated an object detection dataset of more than **2000** cells for a total of **7** categories.
|
| 104 |
+
|
| 105 |
+
| class number | class name |
|
| 106 |
+
| :----------- | :------------------ |
|
| 107 |
+
| 0 | normal_columnar |
|
| 108 |
+
| 1 | normal_intermediate |
|
| 109 |
+
| 2 | normal_superficiel |
|
| 110 |
+
| 3 | carcinoma_in_situ |
|
| 111 |
+
| 4 | light_dysplastic |
|
| 112 |
+
| 5 | moderate_dysplastic |
|
| 113 |
+
| 6 | severe_dysplastic |
|
| 114 |
+
|
| 115 |
+
We decided to share about 800 of them, which should be an adequate number for further test and study.
|
| 116 |
+
|
| 117 |
+
</details>
|
| 118 |
+
|
| 119 |
+
<details open>
|
| 120 |
+
<summary><h4>Train custom models</h4></summary>
|
| 121 |
+
|
| 122 |
+
You can train your own custom model as long as it can work properly.
|
| 123 |
+
|
| 124 |
+
</details>
|
| 125 |
+
|
| 126 |
+
|
| 127 |
+
|
| 128 |
+
</div>
|
| 129 |
+
|
| 130 |
+
### <div align="center"><h2>Training</h2></div>
|
| 131 |
+
|
| 132 |
+
<details open>
|
| 133 |
+
<summary><h4>example weights</h4></summary>
|
| 134 |
+
|
| 135 |
+
Example models of the project are trained with different methods, ranging from Convolutional Neutral Network to Vision Transformer.
|
| 136 |
+
|
| 137 |
+
| Model Name | Training Device | Open Source Repository for Reference | Average AP |
|
| 138 |
+
| ------------ | :--------------------------------: | :--------------------------------------------: | :--------: |
|
| 139 |
+
| yolov5_based | NVIDIA GeForce RTX 4090, 24563.5MB | https://github.com/ultralytics/yolov5.git | 0.721 |
|
| 140 |
+
| yolov8_based | NVIDIA GeForce RTX 4090, 24563.5MB | https://github.com/ultralytics/ultralytics.git | 0.810 |
|
| 141 |
+
| vit_based | NVIDIA GeForce RTX 4090, 24563.5MB | https://github.com/hustvl/YOLOS.git | 0.834 |
|
| 142 |
+
| detr_based | NVIDIA GeForce RTX 4090, 24563.5MB | https://github.com/lyuwenyu/RT-DETR.git | 0.859 |
|
| 143 |
+
|
| 144 |
+
</details>
|
| 145 |
+
|
| 146 |
+
<details open>
|
| 147 |
+
<summary><h4>architecture baselines</h4></summary>
|
| 148 |
+
|
| 149 |
+
- #### **YOLO**
|
| 150 |
+
|
| 151 |
+
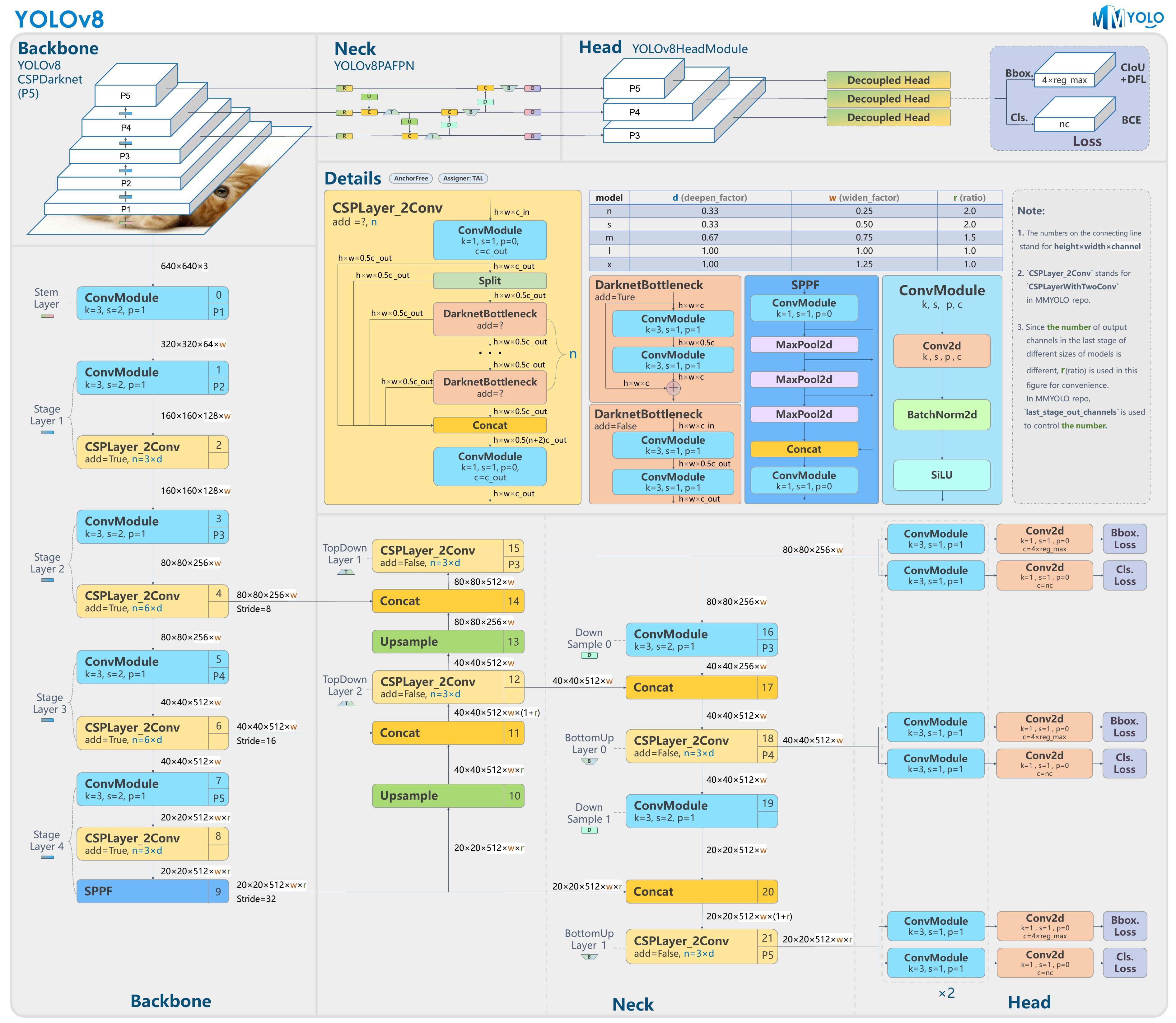
|
| 152 |
+
|
| 153 |
+
|
| 154 |
+
|
| 155 |
+
- #### **Vision Transformer**
|
| 156 |
+
|
| 157 |
+
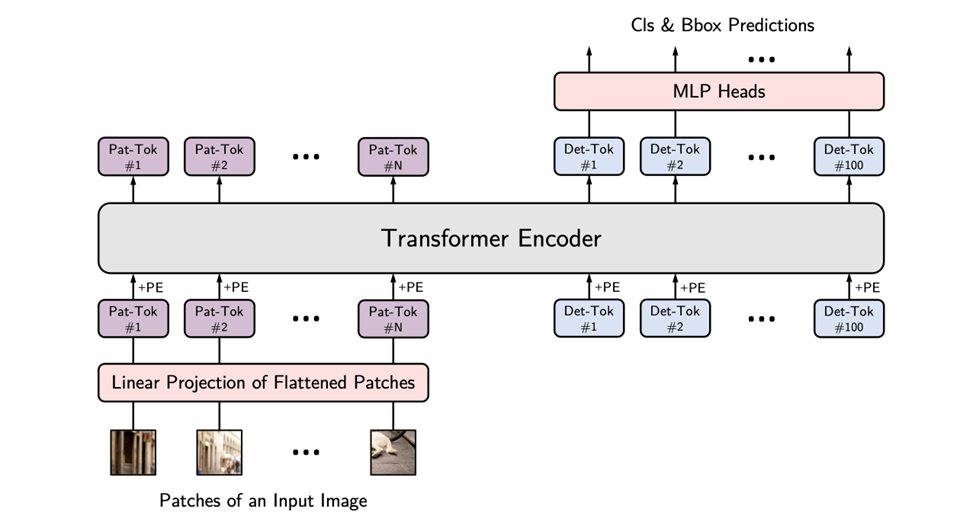
|
| 158 |
+
|
| 159 |
+
- #### **DEtection TRansformer**
|
| 160 |
+
|
| 161 |
+
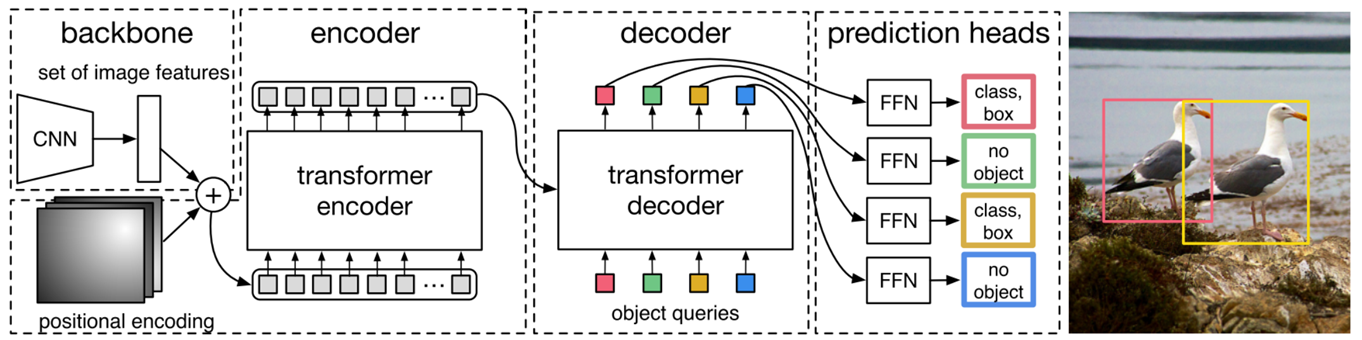
|
| 162 |
+
|
| 163 |
+
</details>
|
| 164 |
+
|
| 165 |
+
|
| 166 |
+
|
| 167 |
+
</div>
|
| 168 |
+
|
| 169 |
+
### <div align="center"><h2>References</h2></div>
|
| 170 |
+
|
| 171 |
+
|
| 172 |
+
|
| 173 |
+
1. Jocher, G., Chaurasia, A., & Qiu, J. (2023). YOLO by Ultralytics (Version 8.0.0) [Computer software]. https://github.com/ultralytics/ultralytics
|
| 174 |
+
|
| 175 |
+
2. [Home - Ultralytics YOLOv8 Docs](https://docs.ultralytics.com/)
|
| 176 |
+
|
| 177 |
+
3. Jocher, G. (2020). YOLOv5 by Ultralytics (Version 7.0) [Computer software]. https://doi.org/10.5281/zenodo.3908559
|
| 178 |
+
|
| 179 |
+
|
| 180 |
+
|
| 181 |
+
4. [GitHub - hustvl/YOLOS: [NeurIPS 2021\] You Only Look at One Sequence](https://github.com/hustvl/YOLOS)
|
| 182 |
+
|
| 183 |
+
5. [GitHub - ViTAE-Transformer/ViTDet: Unofficial implementation for [ECCV'22\] "Exploring Plain Vision Transformer Backbones for Object Detection"](https://github.com/ViTAE-Transformer/ViTDet)
|
| 184 |
+
|
| 185 |
+
6. Touvron, H., Cord, M., Douze, M., Massa, F., Sablayrolles, A., & J'egou, H. (2020). Training data-efficient image transformers & distillation through attention. *International Conference on Machine Learning*.
|
| 186 |
+
|
| 187 |
+
7. Fang, Y., Liao, B., Wang, X., Fang, J., Qi, J., Wu, R., Niu, J., & Liu, W. (2021). You Only Look at One Sequence: Rethinking Transformer in Vision through Object Detection. *Neural Information Processing Systems*.
|
| 188 |
+
|
| 189 |
+
8. [YOLOS (huggingface.co)](https://huggingface.co/docs/transformers/main/en/model_doc/yolos)
|
| 190 |
+
|
| 191 |
+
|
| 192 |
+
|
| 193 |
+
9. Lv, W., Xu, S., Zhao, Y., Wang, G., Wei, J., Cui, C., Du, Y., Dang, Q., & Liu, Y. (2023). DETRs Beat YOLOs on Real-time Object Detection. *ArXiv, abs/2304.08069*.
|
| 194 |
+
|
| 195 |
+
10. [GitHub - facebookresearch/detr: End-to-End Object Detection with Transformers](https://github.com/facebookresearch/detr)
|
| 196 |
+
|
| 197 |
+
11. [PaddleDetection/configs/rtdetr at develop · PaddlePaddle/PaddleDetection · GitHub](https://github.com/PaddlePaddle/PaddleDetection/tree/develop/configs/rtdetr)
|
| 198 |
+
|
| 199 |
+
12. [GitHub - lyuwenyu/RT-DETR: Official RT-DETR (RTDETR paddle pytorch), Real-Time DEtection TRansformer, DETRs Beat YOLOs on Real-time Object Detection. 🔥 🔥 🔥](https://github.com/lyuwenyu/RT-DETR)
|
| 200 |
+
|
| 201 |
+
|
| 202 |
+
|
| 203 |
+
13. J. Hu, L. Shen and G. Sun, "Squeeze-and-Excitation Networks," 2018 IEEE/CVF Conference on Computer Vision and Pattern Recognition, Salt Lake City, UT, USA, 2018, pp. 7132-7141, doi: 10.1109/CVPR.2018.00745.
|
| 204 |
+
|
| 205 |
+
14. Carion, N., Massa, F., Synnaeve, G., Usunier, N., Kirillov, A., & Zagoruyko, S. (2020). End-to-End Object Detection with Transformers. ArXiv, abs/2005.12872.
|
| 206 |
+
|
| 207 |
+
15. Beal, J., Kim, E., Tzeng, E., Park, D., Zhai, A., & Kislyuk, D. (2020). Toward Transformer-Based Object Detection. ArXiv, abs/2012.09958.
|
| 208 |
+
|
| 209 |
+
16. Liu, Z., Lin, Y., Cao, Y., Hu, H., Wei, Y., Zhang, Z., Lin, S., & Guo, B. (2021). Swin Transformer: Hierarchical Vision Transformer using Shifted Windows. 2021 IEEE/CVF International Conference on Computer Vision (ICCV), 9992-10002.
|
| 210 |
+
|
| 211 |
+
17. Zong, Z., Song, G., & Liu, Y. (2022). DETRs with Collaborative Hybrid Assignments Training. ArXiv, abs/2211.12860.
|
| 212 |
+
|
| 213 |
+
|
| 214 |
+
|
| 215 |
+
</div>
|
| 216 |
+
|
| 217 |
+
### <div align="center"><h2>Contact</h2></div>
|
| 218 |
+
|
| 219 |
+
*Feel free to contact me through GitHub issues or directly send me a mail if you have any questions about the project.* 🐼
|
| 220 |
+
|
| 221 |
+
My Gmail Address 👉 [email protected]
|
| 222 |
+
|
app.py
ADDED
|
@@ -0,0 +1,546 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
import argparse
|
| 2 |
+
import csv
|
| 3 |
+
import random
|
| 4 |
+
import sys
|
| 5 |
+
import os
|
| 6 |
+
import wget
|
| 7 |
+
from collections import Counter
|
| 8 |
+
from pathlib import Path
|
| 9 |
+
|
| 10 |
+
import cv2
|
| 11 |
+
import gradio as gr
|
| 12 |
+
import numpy as np
|
| 13 |
+
from matplotlib import font_manager
|
| 14 |
+
from ultralytics import YOLO
|
| 15 |
+
|
| 16 |
+
ROOT_PATH = sys.path[0] # 项目根目录
|
| 17 |
+
|
| 18 |
+
fonts_list = ["SimSun.ttf", "TimesNewRoman.ttf", "malgun.ttf"] # 字体列表
|
| 19 |
+
fonts_directory_path = Path(ROOT_PATH, "fonts")
|
| 20 |
+
|
| 21 |
+
data_url_dict = {
|
| 22 |
+
"SimSun.ttf": "https://raw.githubusercontent.com/Tsumugii24/Typora-images/main/files/SimSun.ttf",
|
| 23 |
+
"TimesNewRoman.ttf": "https://raw.githubusercontent.com/Tsumugii24/Typora-images/main/files/TimesNewRoman.ttf",
|
| 24 |
+
"malgun.ttf": "https://raw.githubusercontent.com/Tsumugii24/Typora-images/main/files/malgun.ttf",
|
| 25 |
+
}
|
| 26 |
+
|
| 27 |
+
|
| 28 |
+
# 判断字体文件是否存在
|
| 29 |
+
def is_fonts(fonts_dir):
|
| 30 |
+
if fonts_dir.is_dir():
|
| 31 |
+
# 如果本地字体库存在
|
| 32 |
+
local_list = os.listdir(fonts_dir) # 本地字体库
|
| 33 |
+
|
| 34 |
+
font_diff = list(set(fonts_list).difference(set(local_list)))
|
| 35 |
+
|
| 36 |
+
if font_diff != []:
|
| 37 |
+
# 缺失字体
|
| 38 |
+
download_fonts(font_diff) # 下载缺失的字体
|
| 39 |
+
else:
|
| 40 |
+
print(f"{fonts_list}[bold green]Required fonts already downloaded![/bold green]")
|
| 41 |
+
else:
|
| 42 |
+
# 本地字体库不存在,创建字体库
|
| 43 |
+
print("[bold red]Local fonts library does not exist, creating now...[/bold red]")
|
| 44 |
+
download_fonts(fonts_list) # 创建字体库
|
| 45 |
+
|
| 46 |
+
# 下载字体
|
| 47 |
+
def download_fonts(font_diff):
|
| 48 |
+
global font_name
|
| 49 |
+
|
| 50 |
+
for k, v in data_url_dict.items():
|
| 51 |
+
if k in font_diff:
|
| 52 |
+
font_name = v.split("/")[-1] # 字体名称
|
| 53 |
+
fonts_directory_path.mkdir(parents=True, exist_ok=True) # 创建目录
|
| 54 |
+
|
| 55 |
+
file_path = f"{ROOT_PATH}/fonts/{font_name}" # 字体路径
|
| 56 |
+
# 下载字体文件
|
| 57 |
+
wget.download(v, file_path)
|
| 58 |
+
|
| 59 |
+
is_fonts(fonts_directory_path)
|
| 60 |
+
|
| 61 |
+
# --------------------- 字体库 ---------------------
|
| 62 |
+
SimSun_path = f"{ROOT_PATH}/fonts/SimSun.ttf" # 宋体文件路径
|
| 63 |
+
TimesNesRoman_path = f"{ROOT_PATH}/fonts/TimesNewRoman.ttf" # 新罗马字体文件路径
|
| 64 |
+
# 宋体
|
| 65 |
+
SimSun = font_manager.FontProperties(fname=SimSun_path, size=12)
|
| 66 |
+
# 新罗马字体
|
| 67 |
+
TimesNesRoman = font_manager.FontProperties(fname=TimesNesRoman_path, size=12)
|
| 68 |
+
|
| 69 |
+
import yaml
|
| 70 |
+
from PIL import Image, ImageDraw, ImageFont
|
| 71 |
+
|
| 72 |
+
# from util.fonts_opt import is_fonts
|
| 73 |
+
|
| 74 |
+
ROOT_PATH = sys.path[0] # 根目录
|
| 75 |
+
|
| 76 |
+
# Gradio version
|
| 77 |
+
GYD_VERSION = "Gradio Lesion-Cells DET v1.0"
|
| 78 |
+
|
| 79 |
+
# 文件后缀
|
| 80 |
+
suffix_list = [".csv", ".yaml"]
|
| 81 |
+
|
| 82 |
+
# 字体大小
|
| 83 |
+
FONTSIZE = 25
|
| 84 |
+
|
| 85 |
+
# 目标尺寸
|
| 86 |
+
obj_style = ["small", "medium", "large"]
|
| 87 |
+
|
| 88 |
+
# title = "Multi-granularity Lesion Cells Object Detection based on deep neural network"
|
| 89 |
+
# description = "<center><h3>Description: This is a WebUI interface demo, Maintained by G1 JIANG SHUFAN</h3></center>"
|
| 90 |
+
|
| 91 |
+
GYD_TITLE = """
|
| 92 |
+
<p align='center'><a href='https://github.com/Tsumugii24/lesion-cells-det'>
|
| 93 |
+
<img src='https://cdn.jsdelivr.net/gh/Tsumugii24/Typora-images@main/images/2023%2F11%2F12%2F2ce6ad153e2e862d5017864fc5087e59-image-20231112230354573-56a688.png' alt='Simple Icons' ></a>
|
| 94 |
+
<center><h1>Multi-granularity Lesion Cells Object Detection based on deep neural network</h1></center>
|
| 95 |
+
<center><h3>Description: This is a WebUI interface demo, Maintained by G1 JIANG SHUFAN</h3></center>
|
| 96 |
+
</p>
|
| 97 |
+
"""
|
| 98 |
+
|
| 99 |
+
GYD_SUB_TITLE = """
|
| 100 |
+
Here is My GitHub Homepage: https://github.com/Tsumugii24 😊
|
| 101 |
+
"""
|
| 102 |
+
|
| 103 |
+
EXAMPLES_DET = [
|
| 104 |
+
["./img_examples/test/moderate0.BMP", "detr_based", "cpu", 640, 0.6,
|
| 105 |
+
0.5, 10, "all range"],
|
| 106 |
+
["./img_examples/test/normal_co0.BMP", "vit_based", "cpu", 640, 0.5,
|
| 107 |
+
0.5, 20, "all range"],
|
| 108 |
+
["./img_examples/test/1280_1920_1.jpg", "yolov8_based", "cpu", 1280, 0.4, 0.5, 15,
|
| 109 |
+
"all range"],
|
| 110 |
+
["./img_examples/test/normal_inter1.BMP", "detr_based", "cpu", 640, 0.4,
|
| 111 |
+
0.5, 30, "all range"],
|
| 112 |
+
["./img_examples/test/1920_1280_1.jpg", "yolov8_based", "cpu", 1280, 0.4, 0.5, 20,
|
| 113 |
+
"all range"],
|
| 114 |
+
["./img_examples/test/severe2.BMP", "detr_based", "cpu", 640, 0.5,
|
| 115 |
+
0.5, 20, "all range"]
|
| 116 |
+
]
|
| 117 |
+
|
| 118 |
+
|
| 119 |
+
def parse_args(known=False):
|
| 120 |
+
parser = argparse.ArgumentParser(description=GYD_VERSION)
|
| 121 |
+
parser.add_argument("--model_name", "-mn", default="detr_based", type=str, help="model name")
|
| 122 |
+
parser.add_argument(
|
| 123 |
+
"--model_cfg",
|
| 124 |
+
"-mc",
|
| 125 |
+
default="./model_config/model_name_cells.yaml",
|
| 126 |
+
type=str,
|
| 127 |
+
help="model config",
|
| 128 |
+
)
|
| 129 |
+
parser.add_argument(
|
| 130 |
+
"--cls_name",
|
| 131 |
+
"-cls",
|
| 132 |
+
default="./cls_name/cls_name_cells_en.yaml",
|
| 133 |
+
type=str,
|
| 134 |
+
help="cls name",
|
| 135 |
+
)
|
| 136 |
+
parser.add_argument(
|
| 137 |
+
"--nms_conf",
|
| 138 |
+
"-conf",
|
| 139 |
+
default=0.5,
|
| 140 |
+
type=float,
|
| 141 |
+
help="model NMS confidence threshold",
|
| 142 |
+
)
|
| 143 |
+
parser.add_argument("--nms_iou", "-iou", default=0.5, type=float, help="model NMS IoU threshold")
|
| 144 |
+
parser.add_argument("--inference_size", "-isz", default=640, type=int, help="model inference size")
|
| 145 |
+
parser.add_argument("--max_detnum", "-mdn", default=50, type=float, help="model max det num")
|
| 146 |
+
parser.add_argument("--slider_step", "-ss", default=0.05, type=float, help="slider step")
|
| 147 |
+
parser.add_argument(
|
| 148 |
+
"--is_login",
|
| 149 |
+
"-isl",
|
| 150 |
+
action="store_true",
|
| 151 |
+
default=False,
|
| 152 |
+
help="is login",
|
| 153 |
+
)
|
| 154 |
+
parser.add_argument('--usr_pwd',
|
| 155 |
+
"-up",
|
| 156 |
+
nargs='+',
|
| 157 |
+
type=str,
|
| 158 |
+
default=["admin", "admin"],
|
| 159 |
+
help="user & password for login")
|
| 160 |
+
parser.add_argument(
|
| 161 |
+
"--is_share",
|
| 162 |
+
"-is",
|
| 163 |
+
action="store_true",
|
| 164 |
+
default=False,
|
| 165 |
+
help="is login",
|
| 166 |
+
)
|
| 167 |
+
parser.add_argument("--server_port", "-sp", default=7860, type=int, help="server port")
|
| 168 |
+
|
| 169 |
+
args = parser.parse_known_args()[0] if known else parser.parse_args()
|
| 170 |
+
return args
|
| 171 |
+
|
| 172 |
+
|
| 173 |
+
# yaml文件解析
|
| 174 |
+
def yaml_parse(file_path):
|
| 175 |
+
return yaml.safe_load(open(file_path, encoding="utf-8").read())
|
| 176 |
+
|
| 177 |
+
|
| 178 |
+
# yaml csv 文件解析
|
| 179 |
+
def yaml_csv(file_path, file_tag):
|
| 180 |
+
file_suffix = Path(file_path).suffix
|
| 181 |
+
if file_suffix == suffix_list[0]:
|
| 182 |
+
# 模型名称
|
| 183 |
+
file_names = [i[0] for i in list(csv.reader(open(file_path)))] # csv版
|
| 184 |
+
elif file_suffix == suffix_list[1]:
|
| 185 |
+
# 模型名称
|
| 186 |
+
file_names = yaml_parse(file_path).get(file_tag) # yaml版
|
| 187 |
+
else:
|
| 188 |
+
print(f"The format of {file_path} is incorrect!")
|
| 189 |
+
sys.exit()
|
| 190 |
+
|
| 191 |
+
return file_names
|
| 192 |
+
|
| 193 |
+
|
| 194 |
+
# 检查网络连接
|
| 195 |
+
def check_online():
|
| 196 |
+
# reference: https://github.com/ultralytics/yolov5/blob/master/utils/general.py
|
| 197 |
+
# check internet connectivity
|
| 198 |
+
import socket
|
| 199 |
+
try:
|
| 200 |
+
socket.create_connection(("1.1.1.1", 443), 5) # check host accessibility
|
| 201 |
+
return True
|
| 202 |
+
except OSError:
|
| 203 |
+
return False
|
| 204 |
+
|
| 205 |
+
|
| 206 |
+
# 标签和边界框颜色设置
|
| 207 |
+
def color_set(cls_num):
|
| 208 |
+
color_list = []
|
| 209 |
+
for i in range(cls_num):
|
| 210 |
+
color = tuple(np.random.choice(range(256), size=3))
|
| 211 |
+
color_list.append(color)
|
| 212 |
+
|
| 213 |
+
return color_list
|
| 214 |
+
|
| 215 |
+
|
| 216 |
+
# 随机生成浅色系或者深色系
|
| 217 |
+
def random_color(cls_num, is_light=True):
|
| 218 |
+
color_list = []
|
| 219 |
+
for i in range(cls_num):
|
| 220 |
+
color = (
|
| 221 |
+
random.randint(0, 127) + int(is_light) * 128,
|
| 222 |
+
random.randint(0, 127) + int(is_light) * 128,
|
| 223 |
+
random.randint(0, 127) + int(is_light) * 128,
|
| 224 |
+
)
|
| 225 |
+
color_list.append(color)
|
| 226 |
+
|
| 227 |
+
return color_list
|
| 228 |
+
|
| 229 |
+
|
| 230 |
+
# 检测绘制
|
| 231 |
+
def pil_draw(img, score_l, bbox_l, cls_l, cls_index_l, textFont, color_list):
|
| 232 |
+
img_pil = ImageDraw.Draw(img)
|
| 233 |
+
id = 0
|
| 234 |
+
|
| 235 |
+
for score, (xmin, ymin, xmax, ymax), label, cls_index in zip(score_l, bbox_l, cls_l, cls_index_l):
|
| 236 |
+
img_pil.rectangle([xmin, ymin, xmax, ymax], fill=None, outline=color_list[cls_index], width=2) # 边界框
|
| 237 |
+
countdown_msg = f"{label} {score:.2f}"
|
| 238 |
+
# text_w, text_h = textFont.getsize(countdown_msg) # 标签尺寸 pillow 9.5.0
|
| 239 |
+
# left, top, left + width, top + height
|
| 240 |
+
# 标签尺寸 pillow 10.0.0
|
| 241 |
+
text_xmin, text_ymin, text_xmax, text_ymax = textFont.getbbox(countdown_msg)
|
| 242 |
+
# 标签背景
|
| 243 |
+
img_pil.rectangle(
|
| 244 |
+
# (xmin, ymin, xmin + text_w, ymin + text_h), # pillow 9.5.0
|
| 245 |
+
(xmin, ymin, xmin + text_xmax - text_xmin, ymin + text_ymax - text_ymin), # pillow 10.0.0
|
| 246 |
+
fill=color_list[cls_index],
|
| 247 |
+
outline=color_list[cls_index],
|
| 248 |
+
)
|
| 249 |
+
|
| 250 |
+
# 标签
|
| 251 |
+
img_pil.multiline_text(
|
| 252 |
+
(xmin, ymin),
|
| 253 |
+
countdown_msg,
|
| 254 |
+
fill=(0, 0, 0),
|
| 255 |
+
font=textFont,
|
| 256 |
+
align="center",
|
| 257 |
+
)
|
| 258 |
+
|
| 259 |
+
id += 1
|
| 260 |
+
|
| 261 |
+
return img
|
| 262 |
+
|
| 263 |
+
|
| 264 |
+
# 绘制多边形
|
| 265 |
+
def polygon_drawing(img_mask, canvas, color_seg):
|
| 266 |
+
# ------- RGB转BGR -------
|
| 267 |
+
color_seg = list(color_seg)
|
| 268 |
+
color_seg[0], color_seg[2] = color_seg[2], color_seg[0]
|
| 269 |
+
color_seg = tuple(color_seg)
|
| 270 |
+
# 定义多边形的顶点
|
| 271 |
+
pts = np.array(img_mask, dtype=np.int32)
|
| 272 |
+
|
| 273 |
+
# 多边形绘制
|
| 274 |
+
cv2.drawContours(canvas, [pts], -1, color_seg, thickness=-1)
|
| 275 |
+
|
| 276 |
+
|
| 277 |
+
# 输出分割结果
|
| 278 |
+
def seg_output(img_path, seg_mask_list, color_list, cls_list):
|
| 279 |
+
img = cv2.imread(img_path)
|
| 280 |
+
img_c = img.copy()
|
| 281 |
+
|
| 282 |
+
# w, h = img.shape[1], img.shape[0]
|
| 283 |
+
|
| 284 |
+
# 获取分割坐标
|
| 285 |
+
for seg_mask, cls_index in zip(seg_mask_list, cls_list):
|
| 286 |
+
img_mask = []
|
| 287 |
+
for i in range(len(seg_mask)):
|
| 288 |
+
# img_mask.append([seg_mask[i][0] * w, seg_mask[i][1] * h])
|
| 289 |
+
img_mask.append([seg_mask[i][0], seg_mask[i][1]])
|
| 290 |
+
|
| 291 |
+
polygon_drawing(img_mask, img_c, color_list[int(cls_index)]) # 绘制分割图形
|
| 292 |
+
|
| 293 |
+
img_mask_merge = cv2.addWeighted(img, 0.3, img_c, 0.7, 0) # 合并图像
|
| 294 |
+
|
| 295 |
+
return img_mask_merge
|
| 296 |
+
|
| 297 |
+
|
| 298 |
+
# 目标检测和图像分割模型加载
|
| 299 |
+
def model_loading(img_path, device_opt, conf, iou, infer_size, max_det, yolo_model="yolov8n.pt"):
|
| 300 |
+
model = YOLO(yolo_model)
|
| 301 |
+
|
| 302 |
+
results = model(source=img_path, device=device_opt, imgsz=infer_size, conf=conf, iou=iou, max_det=max_det)
|
| 303 |
+
results = list(results)[0]
|
| 304 |
+
return results
|
| 305 |
+
|
| 306 |
+
|
| 307 |
+
# YOLOv8图片检测函数
|
| 308 |
+
def yolo_det_img(img_path, model_name, device_opt, infer_size, conf, iou, max_det, obj_size):
|
| 309 |
+
global model, model_name_tmp, device_tmp
|
| 310 |
+
|
| 311 |
+
s_obj, m_obj, l_obj = 0, 0, 0
|
| 312 |
+
|
| 313 |
+
area_obj_all = [] # 目标面积
|
| 314 |
+
|
| 315 |
+
score_det_stat = [] # 置信度统计
|
| 316 |
+
bbox_det_stat = [] # 边界框统计
|
| 317 |
+
cls_det_stat = [] # 类别数量统计
|
| 318 |
+
cls_index_det_stat = [] # 1
|
| 319 |
+
|
| 320 |
+
# 模型加载
|
| 321 |
+
predict_results = model_loading(img_path, device_opt, conf, iou, infer_size, max_det, yolo_model=f"{model_name}.pt")
|
| 322 |
+
# 检测参数
|
| 323 |
+
xyxy_list = predict_results.boxes.xyxy.cpu().numpy().tolist()
|
| 324 |
+
conf_list = predict_results.boxes.conf.cpu().numpy().tolist()
|
| 325 |
+
cls_list = predict_results.boxes.cls.cpu().numpy().tolist()
|
| 326 |
+
|
| 327 |
+
# 颜色列表
|
| 328 |
+
color_list = random_color(len(model_cls_name_cp), True)
|
| 329 |
+
|
| 330 |
+
# 图像分割
|
| 331 |
+
if (model_name[-3:] == "seg"):
|
| 332 |
+
# masks_list = predict_results.masks.xyn
|
| 333 |
+
masks_list = predict_results.masks.xy
|
| 334 |
+
img_mask_merge = seg_output(img_path, masks_list, color_list, cls_list)
|
| 335 |
+
img = Image.fromarray(cv2.cvtColor(img_mask_merge, cv2.COLOR_BGRA2RGBA))
|
| 336 |
+
else:
|
| 337 |
+
img = Image.open(img_path)
|
| 338 |
+
|
| 339 |
+
# 判断检测对象是否为空
|
| 340 |
+
if (xyxy_list != []):
|
| 341 |
+
|
| 342 |
+
# ---------------- 加载字体 ----------------
|
| 343 |
+
yaml_index = cls_name.index(".yaml")
|
| 344 |
+
cls_name_lang = cls_name[yaml_index - 2:yaml_index]
|
| 345 |
+
|
| 346 |
+
if cls_name_lang == "zh":
|
| 347 |
+
# Chinese
|
| 348 |
+
textFont = ImageFont.truetype(str(f"{ROOT_PATH}/fonts/SimSun.ttf"), size=FONTSIZE)
|
| 349 |
+
elif cls_name_lang == "en":
|
| 350 |
+
# English
|
| 351 |
+
textFont = ImageFont.truetype(str(f"{ROOT_PATH}/fonts/TimesNewRoman.ttf"), size=FONTSIZE)
|
| 352 |
+
else:
|
| 353 |
+
# others
|
| 354 |
+
textFont = ImageFont.truetype(str(f"{ROOT_PATH}/fonts/malgun.ttf"), size=FONTSIZE)
|
| 355 |
+
|
| 356 |
+
for i in range(len(xyxy_list)):
|
| 357 |
+
|
| 358 |
+
# ------------ 边框坐标 ------------
|
| 359 |
+
x0 = int(xyxy_list[i][0])
|
| 360 |
+
y0 = int(xyxy_list[i][1])
|
| 361 |
+
x1 = int(xyxy_list[i][2])
|
| 362 |
+
y1 = int(xyxy_list[i][3])
|
| 363 |
+
|
| 364 |
+
# ---------- 加入目标尺寸 ----------
|
| 365 |
+
w_obj = x1 - x0
|
| 366 |
+
h_obj = y1 - y0
|
| 367 |
+
area_obj = w_obj * h_obj # 目标尺寸
|
| 368 |
+
|
| 369 |
+
if (obj_size == "small" and area_obj > 0 and area_obj <= 32 ** 2):
|
| 370 |
+
obj_cls_index = int(cls_list[i]) # 类别索引
|
| 371 |
+
cls_index_det_stat.append(obj_cls_index)
|
| 372 |
+
|
| 373 |
+
obj_cls = model_cls_name_cp[obj_cls_index] # 类别
|
| 374 |
+
cls_det_stat.append(obj_cls)
|
| 375 |
+
|
| 376 |
+
bbox_det_stat.append((x0, y0, x1, y1))
|
| 377 |
+
|
| 378 |
+
conf = float(conf_list[i]) # 置信度
|
| 379 |
+
score_det_stat.append(conf)
|
| 380 |
+
|
| 381 |
+
area_obj_all.append(area_obj)
|
| 382 |
+
elif (obj_size == "medium" and area_obj > 32 ** 2 and area_obj <= 96 ** 2):
|
| 383 |
+
obj_cls_index = int(cls_list[i]) # 类别索引
|
| 384 |
+
cls_index_det_stat.append(obj_cls_index)
|
| 385 |
+
|
| 386 |
+
obj_cls = model_cls_name_cp[obj_cls_index] # 类别
|
| 387 |
+
cls_det_stat.append(obj_cls)
|
| 388 |
+
|
| 389 |
+
bbox_det_stat.append((x0, y0, x1, y1))
|
| 390 |
+
|
| 391 |
+
conf = float(conf_list[i]) # 置信度
|
| 392 |
+
score_det_stat.append(conf)
|
| 393 |
+
|
| 394 |
+
area_obj_all.append(area_obj)
|
| 395 |
+
elif (obj_size == "large" and area_obj > 96 ** 2):
|
| 396 |
+
obj_cls_index = int(cls_list[i]) # 类别索引
|
| 397 |
+
cls_index_det_stat.append(obj_cls_index)
|
| 398 |
+
|
| 399 |
+
obj_cls = model_cls_name_cp[obj_cls_index] # 类别
|
| 400 |
+
cls_det_stat.append(obj_cls)
|
| 401 |
+
|
| 402 |
+
bbox_det_stat.append((x0, y0, x1, y1))
|
| 403 |
+
|
| 404 |
+
conf = float(conf_list[i]) # 置信度
|
| 405 |
+
score_det_stat.append(conf)
|
| 406 |
+
|
| 407 |
+
area_obj_all.append(area_obj)
|
| 408 |
+
elif (obj_size == "all range"):
|
| 409 |
+
obj_cls_index = int(cls_list[i]) # 类别索引
|
| 410 |
+
cls_index_det_stat.append(obj_cls_index)
|
| 411 |
+
|
| 412 |
+
obj_cls = model_cls_name_cp[obj_cls_index] # 类别
|
| 413 |
+
cls_det_stat.append(obj_cls)
|
| 414 |
+
|
| 415 |
+
bbox_det_stat.append((x0, y0, x1, y1))
|
| 416 |
+
|
| 417 |
+
conf = float(conf_list[i]) # 置信度
|
| 418 |
+
score_det_stat.append(conf)
|
| 419 |
+
|
| 420 |
+
area_obj_all.append(area_obj)
|
| 421 |
+
|
| 422 |
+
det_img = pil_draw(img, score_det_stat, bbox_det_stat, cls_det_stat, cls_index_det_stat, textFont, color_list)
|
| 423 |
+
|
| 424 |
+
# -------------- 目标尺寸计算 --------------
|
| 425 |
+
for i in range(len(area_obj_all)):
|
| 426 |
+
if (0 < area_obj_all[i] <= 32 ** 2):
|
| 427 |
+
s_obj = s_obj + 1
|
| 428 |
+
elif (32 ** 2 < area_obj_all[i] <= 96 ** 2):
|
| 429 |
+
m_obj = m_obj + 1
|
| 430 |
+
elif (area_obj_all[i] > 96 ** 2):
|
| 431 |
+
l_obj = l_obj + 1
|
| 432 |
+
|
| 433 |
+
sml_obj_total = s_obj + m_obj + l_obj
|
| 434 |
+
objSize_dict = {obj_style[i]: [s_obj, m_obj, l_obj][i] / sml_obj_total for i in range(3)}
|
| 435 |
+
|
| 436 |
+
# ------------ 类别统计 ------------
|
| 437 |
+
clsRatio_dict = {}
|
| 438 |
+
clsDet_dict = Counter(cls_det_stat)
|
| 439 |
+
clsDet_dict_sum = sum(clsDet_dict.values())
|
| 440 |
+
for k, v in clsDet_dict.items():
|
| 441 |
+
clsRatio_dict[k] = v / clsDet_dict_sum
|
| 442 |
+
|
| 443 |
+
gr.Info("Inference Success!")
|
| 444 |
+
return det_img, objSize_dict, clsRatio_dict
|
| 445 |
+
else:
|
| 446 |
+
raise gr.Error("Failed! This model cannot detect anything from this image, Please try another one.")
|
| 447 |
+
|
| 448 |
+
|
| 449 |
+
def main(args):
|
| 450 |
+
gr.close_all()
|
| 451 |
+
|
| 452 |
+
global model_cls_name_cp, cls_name
|
| 453 |
+
|
| 454 |
+
nms_conf = args.nms_conf
|
| 455 |
+
nms_iou = args.nms_iou
|
| 456 |
+
model_name = args.model_name
|
| 457 |
+
model_cfg = args.model_cfg
|
| 458 |
+
cls_name = args.cls_name
|
| 459 |
+
inference_size = args.inference_size
|
| 460 |
+
max_detnum = args.max_detnum
|
| 461 |
+
slider_step = args.slider_step
|
| 462 |
+
|
| 463 |
+
# is_fonts(f"{ROOT_PATH}/fonts") # 检查字体文件
|
| 464 |
+
|
| 465 |
+
model_names = yaml_csv(model_cfg, "model_names") # 模型名称
|
| 466 |
+
model_cls_name = yaml_csv(cls_name, "model_cls_name") # 类别名称
|
| 467 |
+
|
| 468 |
+
model_cls_name_cp = model_cls_name.copy() # 类别名称
|
| 469 |
+
|
| 470 |
+
custom_theme = gr.themes.Soft(primary_hue="slate", secondary_hue="sky").set(
|
| 471 |
+
button_secondary_background_fill="*neutral_100",
|
| 472 |
+
button_secondary_background_fill_hover="*neutral_200")
|
| 473 |
+
custom_css = '''#disp_image {
|
| 474 |
+
text-align: center; /* Horizontally center the content */
|
| 475 |
+
}'''
|
| 476 |
+
|
| 477 |
+
# ------------ Gradio Blocks ------------
|
| 478 |
+
with gr.Blocks(theme=custom_theme, css=custom_css) as gyd:
|
| 479 |
+
with gr.Row():
|
| 480 |
+
gr.Markdown(GYD_TITLE)
|
| 481 |
+
with gr.Row():
|
| 482 |
+
gr.Markdown(GYD_SUB_TITLE)
|
| 483 |
+
with gr.Row():
|
| 484 |
+
with gr.Column(scale=1):
|
| 485 |
+
with gr.Tabs():
|
| 486 |
+
with gr.TabItem("Object Detection"):
|
| 487 |
+
with gr.Row():
|
| 488 |
+
inputs_img = gr.Image(image_mode="RGB", type="filepath", label="original image")
|
| 489 |
+
with gr.Row():
|
| 490 |
+
# device_opt = gr.Radio(choices=["cpu", "0", "1", "2", "3"], value="cpu", label="device")
|
| 491 |
+
device_opt = gr.Radio(choices=["cpu", "gpu 0", "gpu 1", "gpu 2", "gpu 3"], value="cpu",
|
| 492 |
+
label="device")
|
| 493 |
+
with gr.Row():
|
| 494 |
+
inputs_model = gr.Dropdown(choices=model_names, value=model_name, type="value",
|
| 495 |
+
label="model")
|
| 496 |
+
with gr.Row():
|
| 497 |
+
inputs_size = gr.Slider(320, 1600, step=1, value=inference_size, label="inference size")
|
| 498 |
+
max_det = gr.Slider(1, 100, step=1, value=max_detnum, label="max bbox number")
|
| 499 |
+
with gr.Row():
|
| 500 |
+
input_conf = gr.Slider(0, 1, step=slider_step, value=nms_conf, label="confidence threshold")
|
| 501 |
+
inputs_iou = gr.Slider(0, 1, step=slider_step, value=nms_iou, label="IoU threshold")
|
| 502 |
+
with gr.Row():
|
| 503 |
+
obj_size = gr.Radio(choices=["all range", "small", "medium", "large"], value="all range",
|
| 504 |
+
label="cell size(relative)")
|
| 505 |
+
with gr.Row():
|
| 506 |
+
gr.ClearButton(inputs_img, value="clear")
|
| 507 |
+
det_btn_img = gr.Button(value='submit', variant="primary")
|
| 508 |
+
with gr.Row():
|
| 509 |
+
gr.Examples(examples=EXAMPLES_DET,
|
| 510 |
+
fn=yolo_det_img,
|
| 511 |
+
inputs=[inputs_img, inputs_model, device_opt, inputs_size, input_conf,
|
| 512 |
+
inputs_iou, max_det, obj_size],
|
| 513 |
+
# outputs=[outputs_img, outputs_objSize, outputs_clsSize],
|
| 514 |
+
cache_examples=False)
|
| 515 |
+
|
| 516 |
+
with gr.Column(scale=1):
|
| 517 |
+
with gr.Tabs():
|
| 518 |
+
with gr.TabItem("Object Detection"):
|
| 519 |
+
with gr.Row():
|
| 520 |
+
outputs_img = gr.Image(type="pil", label="detection results")
|
| 521 |
+
with gr.Row():
|
| 522 |
+
outputs_objSize = gr.Label(label="Percentage Statistics of cells size(relative)")
|
| 523 |
+
with gr.Row():
|
| 524 |
+
outputs_clsSize = gr.Label(label="Percentage Statistics of cells lesion degree")
|
| 525 |
+
|
| 526 |
+
det_btn_img.click(fn=yolo_det_img,
|
| 527 |
+
inputs=[
|
| 528 |
+
inputs_img, inputs_model, device_opt, inputs_size, input_conf, inputs_iou, max_det,
|
| 529 |
+
obj_size],
|
| 530 |
+
outputs=[outputs_img, outputs_objSize, outputs_clsSize])
|
| 531 |
+
|
| 532 |
+
return gyd
|
| 533 |
+
|
| 534 |
+
|
| 535 |
+
if __name__ == "__main__":
|
| 536 |
+
args = parse_args()
|
| 537 |
+
gyd = main(args)
|
| 538 |
+
is_share = args.is_share
|
| 539 |
+
|
| 540 |
+
gyd.queue().launch(
|
| 541 |
+
inbrowser=True, # 自动打开默认浏览器
|
| 542 |
+
share=is_share, # 项目共享,其他设备可以访问
|
| 543 |
+
favicon_path="favicons/logo.ico", # 网页图标
|
| 544 |
+
show_error=True, # 在浏览器控制台中显示错误信息
|
| 545 |
+
quiet=True, # 禁止大多数打印语句
|
| 546 |
+
)
|
cls_name/cls_name_cells_en.yaml
ADDED
|
@@ -0,0 +1 @@
|
|
|
|
|
|
|
| 1 |
+
model_cls_name: ['severe_dysplastic', "carcinoma_in_situ", "light_dysplastic", "moderate_dysplastic", "normal_columnar", "normal_intermediate", "normal_superficiel"]
|
cls_name/cls_name_cells_zh.yaml
ADDED
|
@@ -0,0 +1 @@
|
|
|
|
|
|
|
| 1 |
+
model_cls_name: ['严重发育不良', "癌变细胞", "轻度发育不良", "中度发育不良", "正常柱状细胞", "正常中间体细胞", "正常表皮细胞"]
|
favicons/Lesion-Cells DET.png
ADDED
|
|
favicons/logo.ico
ADDED
|
|
img_examples/ground_truth_label/carcinoma_in_situ/149315671-149315740-001.BMP
ADDED
|
|
img_examples/ground_truth_label/carcinoma_in_situ/149315671-149315740-002.BMP
ADDED
|
|
img_examples/ground_truth_label/carcinoma_in_situ/149315671-149315740-003.BMP
ADDED
|
|
img_examples/ground_truth_label/carcinoma_in_situ/149315671-149315740-004.BMP
ADDED
|
|
img_examples/ground_truth_label/carcinoma_in_situ/149315671-149315749-001.BMP
ADDED
|
|
img_examples/ground_truth_label/carcinoma_in_situ/149315671-149315749-002.BMP
ADDED
|
|
img_examples/ground_truth_label/carcinoma_in_situ/149315775-149315790-001.BMP
ADDED
|
|
img_examples/ground_truth_label/carcinoma_in_situ/149315775-149315790-002.BMP
ADDED
|
|
img_examples/ground_truth_label/carcinoma_in_situ/149315775-149315790-003.BMP
ADDED
|
|
img_examples/ground_truth_label/carcinoma_in_situ/149316025-149316032-001.BMP
ADDED
|
|
img_examples/ground_truth_label/carcinoma_in_situ/149316117-149316122-002.BMP
ADDED
|
|
img_examples/ground_truth_label/carcinoma_in_situ/149316117-149316122-003.BMP
ADDED
|
|
img_examples/ground_truth_label/carcinoma_in_situ/149316117-149316122-004.BMP
ADDED
|
|
img_examples/ground_truth_label/carcinoma_in_situ/149316117-149316131-001.BMP
ADDED
|
|
img_examples/ground_truth_label/carcinoma_in_situ/149316117-149316131-002.BMP
ADDED
|
|
img_examples/ground_truth_label/carcinoma_in_situ/149316426-149316462-001.BMP
ADDED
|
|
img_examples/ground_truth_label/carcinoma_in_situ/149316426-149316462-002.BMP
ADDED
|
|
img_examples/ground_truth_label/carcinoma_in_situ/149316426-149316462-003.BMP
ADDED
|
|
img_examples/ground_truth_label/carcinoma_in_situ/149316426-149316462-004.BMP
ADDED
|
|
img_examples/ground_truth_label/carcinoma_in_situ/149317114-149317152-001.BMP
ADDED
|
|
img_examples/ground_truth_label/carcinoma_in_situ/149317114-149317152-002.BMP
ADDED
|
|
img_examples/ground_truth_label/carcinoma_in_situ/149317114-149317152-004.BMP
ADDED
|
|
img_examples/ground_truth_label/carcinoma_in_situ/149357956-149358043-001.BMP
ADDED
|
|
img_examples/ground_truth_label/carcinoma_in_situ/153826597-153826619-001.BMP
ADDED
|
|
img_examples/ground_truth_label/carcinoma_in_situ/153826597-153826619-002.BMP
ADDED
|
|
img_examples/ground_truth_label/carcinoma_in_situ/153827595-153827657-001.BMP
ADDED
|
|
img_examples/ground_truth_label/carcinoma_in_situ/153827595-153827657-002.BMP
ADDED
|
|
img_examples/ground_truth_label/carcinoma_in_situ/153827595-153827664-001.BMP
ADDED
|
|
img_examples/ground_truth_label/carcinoma_in_situ/153827595-153827669-001.BMP
ADDED
|
|
img_examples/ground_truth_label/carcinoma_in_situ/153830680-153830693-001.BMP
ADDED
|
|
img_examples/ground_truth_label/carcinoma_in_situ/153831027-153831036-001.BMP
ADDED
|
|
img_examples/ground_truth_label/carcinoma_in_situ/153831027-153831036-002.BMP
ADDED
|
|
img_examples/ground_truth_label/carcinoma_in_situ/153831027-153831045-001.BMP
ADDED
|
|
img_examples/ground_truth_label/carcinoma_in_situ/153831027-153831045-002.BMP
ADDED
|
|
img_examples/ground_truth_label/carcinoma_in_situ/153831027-153831045-003.BMP
ADDED
|
|
img_examples/ground_truth_label/carcinoma_in_situ/153831426-153831438-001.BMP
ADDED
|
|
img_examples/ground_truth_label/carcinoma_in_situ/153831471-153831479-001.BMP
ADDED
|
|
img_examples/ground_truth_label/carcinoma_in_situ/153831471-153831486-001.BMP
ADDED
|
|
img_examples/ground_truth_label/carcinoma_in_situ/153831471-153831486-002.BMP
ADDED
|
|
img_examples/ground_truth_label/carcinoma_in_situ/153831471-153831486-003.BMP
ADDED
|
|
img_examples/ground_truth_label/carcinoma_in_situ/153831471-153831486-004.BMP
ADDED
|
|
img_examples/ground_truth_label/carcinoma_in_situ/153916114-153916150-001.BMP
ADDED
|
|