---
license: mit
datasets:
- colabfit/MD22_buckyball_catcher
- colabfit/MD22_AT_AT
- colabfit/MD22_stachyose
- colabfit/MD22_AT_AT_CG_CG
- colabfit/MD22_Ac_Ala3_NHMe
- colabfit/MD22_DHA
- colabfit/MD22_double_walled_nanotube
- yairschiff/qm9
- maomlab/Molecule3D
metrics:
- mae
tags:
- equivariant
- graph neural network
- molecular property prediction
---
# GotenNet: Rethinking Efficient 3D Equivariant Graph Neural Networks
[](https://openreview.net/pdf?id=5wxCQDtbMo)
[](https://www.sarpaykent.com/publications/gotennet/)
[](LICENSE)
[](https://pypi.org/project/gotennet/)
[](https://pytorch.org/)
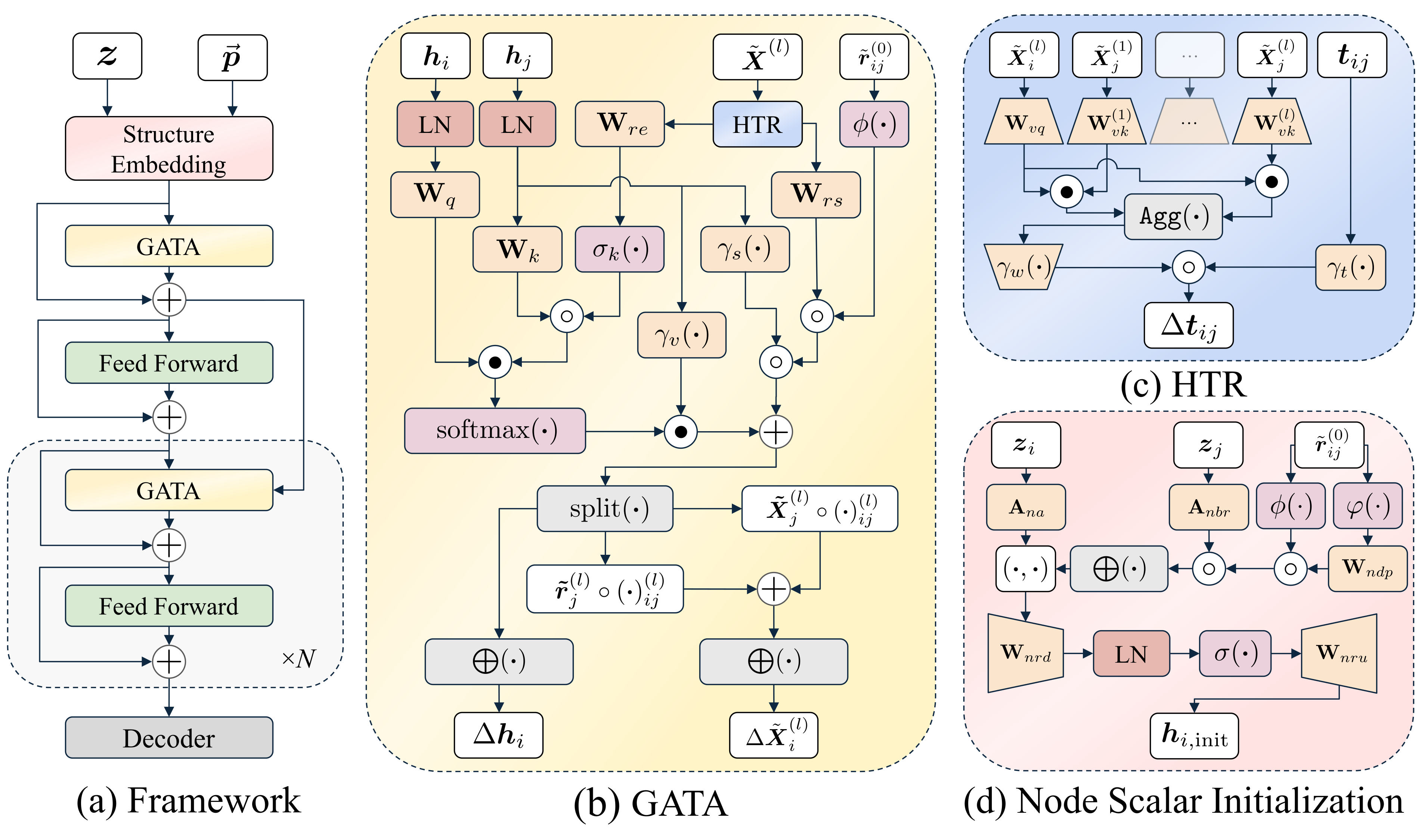
## Overview
This is the official implementation of **"GotenNet: Rethinking Efficient 3D Equivariant Graph Neural Networks"** published at ICLR 2025.
GotenNet introduces a novel framework for modeling 3D molecular structures that achieves state-of-the-art performance while maintaining computational efficiency. Our approach balances expressiveness and efficiency through innovative tensor-based representations and attention mechanisms.
## Table of Contents
- [✨ Key Features](#-key-features)
- [🚀 Installation](#-installation)
- [📦 From PyPI (Recommended)](#-from-pypi-recommended)
- [🔧 From Source](#🔧-from-source)
- [🔬 Usage](#🔬-usage)
- [Using the Model](#using-the-model)
- [Loading Pre-trained Models Programmatically](#loading-pre-trained-models-programmatically)
- [Training a Model](#training-a-model)
- [Testing a Model](#testing-a-model)
- [Configuration](#configuration)
- [🤝 Contributing](#-contributing)
- [📚 Citation](#-citation)
- [📄 License](#-license)
- [Acknowledgements](#acknowledgements)
## ✨ Key Features
- 🔄 **Effective Geometric Tensor Representations**: Leverages geometric tensors without relying on irreducible representations or Clebsch-Gordan transforms
- 🧩 **Unified Structural Embedding**: Introduces geometry-aware tensor attention for improved molecular representation
- 📊 **Hierarchical Tensor Refinement**: Implements a flexible and efficient representation scheme
- 🏆 **State-of-the-Art Performance**: Achieves superior results on QM9, rMD17, MD22, and Molecule3D datasets
- 📈 **Load Pre-trained Models**: Easily load and use pre-trained model checkpoints by name, URL, or local path, with automatic download capabilities.
## 🚀 Installation
### 📦 From PyPI (Recommended)
You can install it using pip:
* **Core Model Only:** Installs only the essential dependencies required to use the `GotenNet` model.
```bash
pip install gotennet
```
* **Full Installation (Core + Training/Utilities):** Installs core dependencies plus libraries needed for training, data handling, logging, etc.
```bash
pip install gotennet[full]
```
### 🔧 From Source
1. **Clone the repository:**
```bash
git clone https://github.com/sarpaykent/gotennet.git
cd gotennet
```
2. **Create and activate a virtual environment** (using conda or venv/uv):
```bash
# Using conda
conda create -n gotennet python=3.10
conda activate gotennet
# Or using venv/uv
uv venv --python 3.10
source .venv/bin/activate
```
3. **Install the package:**
Choose the installation type based on your needs:
* **Core Model Only:** Installs only the essential dependencies required to use the `GotenNet` model.
```bash
pip install .
```
* **Full Installation (Core + Training/Utilities):** Installs core dependencies plus libraries needed for training, data handling, logging, etc.
```bash
pip install .[full]
# Or for editable install:
# pip install -e .[full]
```
*(Note: `uv` can be used as a faster alternative to `pip` for installation, e.g., `uv pip install .[full]`)*
## 🔬 Usage
### Using the Model
Once installed, you can import and use the `GotenNet` model directly in your Python code:
```python
from gotennet import GotenNet
# --- Using the base GotenNet model ---
# Requires manual calculation of edge_index, edge_diff, edge_vec
# Example instantiation
model = GotenNet(
n_atom_basis=256,
n_interactions=4,
# resf of the parameters
)
# Encoded representations can be computed with
h, X = model(atomic_numbers, edge_index, edge_diff, edge_vec)
# --- Using GotenNetWrapper (handles distance calculation) ---
# Expects a PyTorch Geometric Data object or similar dict
# with keys like 'z' (atomic_numbers), 'pos' (positions), 'batch'
# Example instantiation
from gotennet import GotenNetWrapper
wrapped_model = GotenNetWrapper(
n_atom_basis=256,
n_interactions=4,
# rest of the parameters
)
# Encoded representations can be computed with
h, X = wrapped_model(data)
```
### Loading Pre-trained Models Programmatically
You can easily load pre-trained `GotenModel` instances programmatically using the `from_pretrained` class method. This method can accept a model alias (which will be resolved to a download URL), a direct HTTPS URL to a checkpoint file, or a local file path. It handles automatic downloading and caching of checkpoints. Pre-trained model weights and aliases are hosted on the [GotenNet Hugging Face Model Hub](https://huggingface.co/sarpaykent/GotenNet).
```python
from gotennet.models import GotenModel
# Example 1: Load by model alias
# This will automatically download from a known location if not found locally.
# The format is {dataset}_{size}_{target}
model_by_alias = GotenModel.from_pretrained("QM9_small_homo")
# Example 2: Load from a direct URL
model_url = "https://huggingface.co/sarpaykent/GotenNet/resolve/main/pretrained/qm9/small/gotennet_homo.ckpt" # Replace with an actual URL
model_by_url = GotenModel.from_pretrained(model_url)
# Example 3: Load from a local file path
local_model_path = "/path/to/your/local_model.ckpt"
model_by_path = GotenModel.from_pretrained(local_model_path)
# After loading, the model is ready for inference:
predictions = model_by_alias(data_input)
```
For more advanced scenarios, if you only need to load the base `GotenNet` representation module from a local checkpoint (e.g., a checkpoint that only contains representation weights), you can use:
```python
from gotennet.models.representation import GotenNet, GotenNetWrapper
# Example: Load a GotenNet representation from a local file
representation_checkpoint_path = "/path/to/your/local_model.ckpt"
gotennet_model = GotenNet.load_from_checkpoint(representation_checkpoint_path)
# or
gotennet_wrapped = GotenNetWrapper.load_from_checkpoint(representation_checkpoint_path)
```
### Training a Model
After installation, you can use the `train_gotennet` command:
```bash
train_gotennet
```
Or you can run the training script directly:
```bash
python gotennet/scripts/train.py
```
Both methods use Hydra for configuration. You can reproduce U0 target prediction on the QM9 dataset with the following command:
```bash
train_gotennet experiment=qm9_u0.yaml
```
### Testing a Model
To evaluate a trained model, you can use the `test_gotennet` script. When you provide a checkpoint, the script can infer necessary configurations (like dataset and task details) directly from the checkpoint file. This script leverages the `GotenModel.from_pretrained` capabilities, allowing you to specify the model to test by its alias, a direct URL, or a local file path, handling automatic downloads.
Here's how you can use it:
```bash
# Option 1: Test by model alias (e.g., QM9_small_homo)
# The script will automatically download the checkpoint and infer configurations.
test_gotennet checkpoint=QM9_small_homo
# Option 2: Test with a direct checkpoint URL
# The script will automatically download the checkpoint and infer configurations.
test_gotennet checkpoint=https://huggingface.co/sarpaykent/GotenNet/resolve/main/pretrained/qm9/small/gotennet_homo.ckpt
# Option 3: Test with a local checkpoint file path
test_gotennet checkpoint=/path/to/your/local_model.ckpt
```
The script uses [Hydra](https://hydra.cc/) for any additional or overriding configurations if needed, but for straightforward evaluation of a checkpoint, only the `checkpoint` argument is typically required.
### Configuration
The project uses [Hydra](https://hydra.cc/) for configuration management. Configuration files are located in the `configs/` directory.
Main configuration categories:
- `datamodule`: Dataset configurations (md17, qm9, etc.)
- `model`: Model configurations
- `trainer`: Training parameters
- `callbacks`: Callback configurations
- `logger`: Logging configurations
## 🤝 Contributing
We welcome contributions to GotenNet! Please feel free to submit a Pull Request.
## 📚 Citation
Please consider citing our work below if this project is helpful:
```bibtex
@inproceedings{aykent2025gotennet,
author = {Aykent, Sarp and Xia, Tian},
booktitle = {The Thirteenth International Conference on LearningRepresentations},
year = {2025},
title = {{GotenNet: Rethinking Efficient 3D Equivariant Graph Neural Networks}},
url = {https://openreview.net/forum?id=5wxCQDtbMo},
howpublished = {https://openreview.net/forum?id=5wxCQDtbMo},
}
```
## 📄 License
This project is licensed under the MIT License - see the [LICENSE](LICENSE) file for details.
## Acknowledgements
GotenNet is proudly built on the innovative foundations provided by the projects below.
- [e3nn](https://github.com/e3nn/e3nn)
- [PyG](https://github.com/pyg-team/pytorch_geometric)
- [PyTorch Lightning](https://github.com/Lightning-AI/pytorch-lightning)

