Multi-Objective-Guided Discrete Flow Matching for Controllable Biological Sequence Design
Abstract
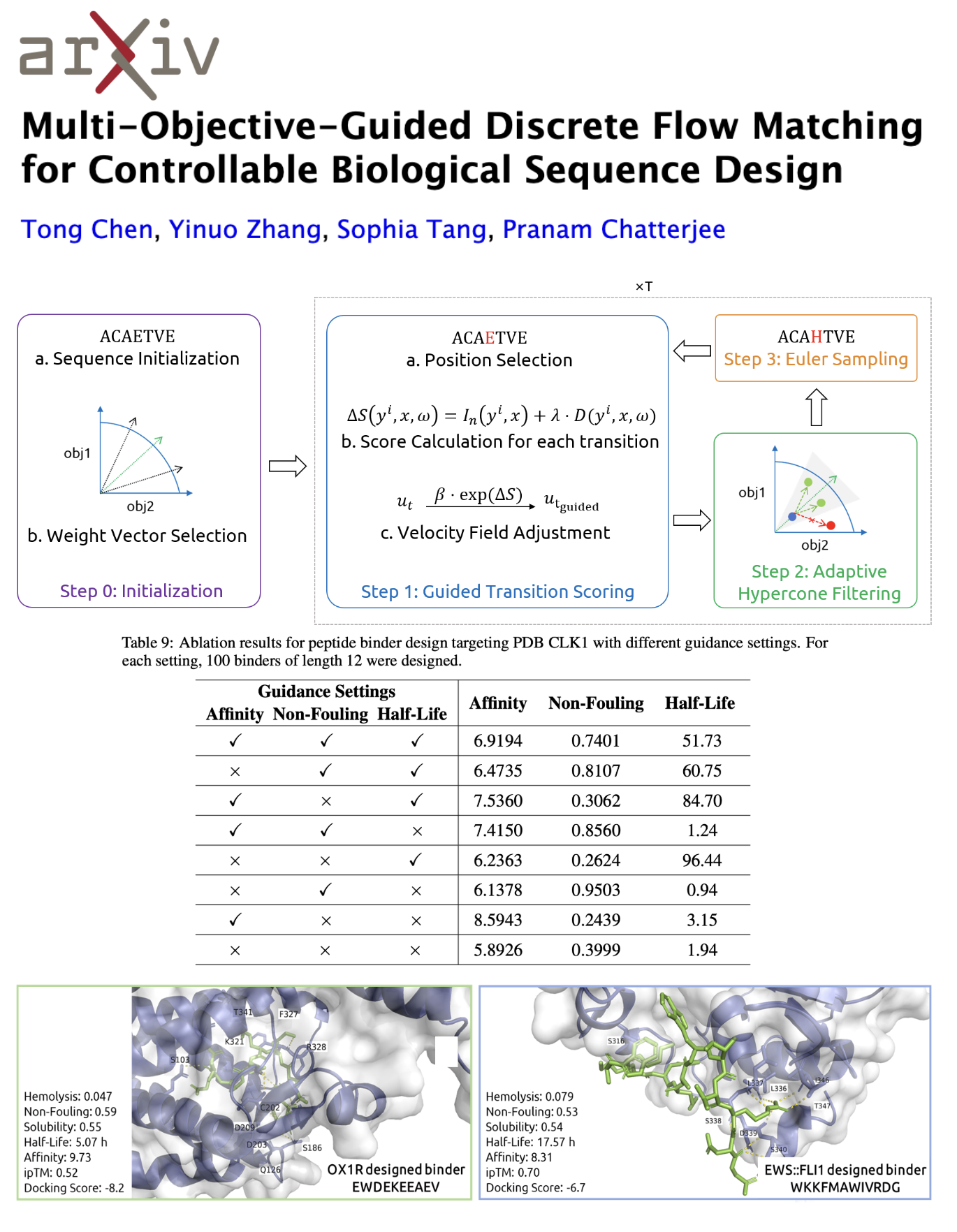
Designing biological sequences that satisfy multiple, often conflicting, functional and biophysical criteria remains a central challenge in biomolecule engineering. While discrete flow matching models have recently shown promise for efficient sampling in high-dimensional sequence spaces, existing approaches address only single objectives or require continuous embeddings that can distort discrete distributions. We present Multi-Objective-Guided Discrete Flow Matching (MOG-DFM), a general framework to steer any pretrained discrete-time flow matching generator toward Pareto-efficient trade-offs across multiple scalar objectives. At each sampling step, MOG-DFM computes a hybrid rank-directional score for candidate transitions and applies an adaptive hypercone filter to enforce consistent multi-objective progression. We also trained two unconditional discrete flow matching models, PepDFM for diverse peptide generation and EnhancerDFM for functional enhancer DNA generation, as base generation models for MOG-DFM. We demonstrate MOG-DFM's effectiveness in generating peptide binders optimized across five properties (hemolysis, non-fouling, solubility, half-life, and binding affinity), and in designing DNA sequences with specific enhancer classes and DNA shapes. In total, MOG-DFM proves to be a powerful tool for multi-property-guided biomolecule sequence design.
Community
This is an automated message from the Librarian Bot. I found the following papers similar to this paper.
The following papers were recommended by the Semantic Scholar API
- Gumbel-Softmax Flow Matching with Straight-Through Guidance for Controllable Biological Sequence Generation (2025)
- ProtFlow: Fast Protein Sequence Design via Flow Matching on Compressed Protein Language Model Embeddings (2025)
- Scoring-Assisted Generative Exploration for Proteins (SAGE-Prot): A Framework for Multi-Objective Protein Optimization via Iterative Sequence Generation and Evaluation (2025)
- CMADiff: Cross-Modal Aligned Diffusion for Controllable Protein Generation (2025)
- IgCraft: A versatile sequence generation framework for antibody discovery and engineering (2025)
- Guide your favorite protein sequence generative model (2025)
- MetaMolGen: A Neural Graph Motif Generation Model for De Novo Molecular Design (2025)
Please give a thumbs up to this comment if you found it helpful!
If you want recommendations for any Paper on Hugging Face checkout this Space
You can directly ask Librarian Bot for paper recommendations by tagging it in a comment:
@librarian-bot
recommend
Models citing this paper 1
Datasets citing this paper 0
No dataset linking this paper
Spaces citing this paper 0
No Space linking this paper