Update README.md
Browse files
README.md
CHANGED
|
@@ -1,21 +1,43 @@
|
|
| 1 |
---
|
| 2 |
license: apache-2.0
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 3 |
---
|
| 4 |
-
|
|
|
|
| 5 |
|
| 6 |
This repository provides PyTorch source code associated with our publication, "A Mamba-Based Foundation Model for Chemistry".
|
| 7 |
|
| 8 |
-
**Paper:** [Arxiv Link]()
|
| 9 |
|
| 10 |
-
**HuggingFace:** [HuggingFace Link](https://huggingface.co/ibm/materials.
|
| 11 |
|
| 12 |
For more information contact: [email protected] or [email protected].
|
| 13 |
|
| 14 |
-
 - MoLMamba
|
| 20 |
|
| 21 |
This repository provides PyTorch source code associated with our publication, "A Mamba-Based Foundation Model for Chemistry".
|
| 22 |
|
| 23 |
+
**Paper:** [Arxiv Link](https://openreview.net/pdf?id=HTgCs0KSTl)
|
| 24 |
|
| 25 |
+
**HuggingFace:** [HuggingFace Link](https://huggingface.co/ibm/materials.smi_ssed)
|
| 26 |
|
| 27 |
For more information contact: [email protected] or [email protected].
|
| 28 |
|
| 29 |
+
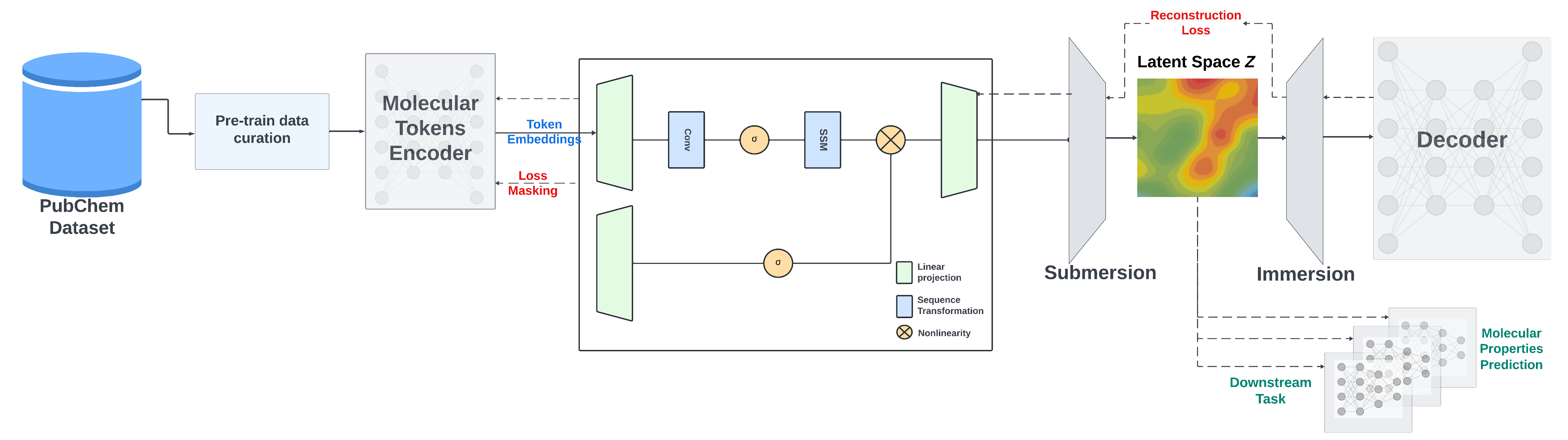
|
| 30 |
|
| 31 |
## Introduction
|
| 32 |
|
| 33 |
+
We present a Mamba-based encoder-decoder chemical foundation model, SMILES-based State-Space Encoder-Decoder (SMI-SSED), pre-trained on a curated dataset of 91 million SMILES samples sourced from PubChem, equivalent to 4 billion molecular tokens. SMI-SSED supports various complex tasks, including quantum property prediction, with two main variants ($336$ and $8 \times 336M$). Our experiments across multiple benchmark datasets demonstrate state-of-the-art performance for various tasks.
|
| 34 |
+
|
| 35 |
+
We provide the model weights in two formats:
|
| 36 |
+
|
| 37 |
+
- PyTorch (`.pt`): [smi_ssed_130.pt](smi_ssed_130.pt)
|
| 38 |
+
- safetensors (`.bin`): [smi_ssed_130.bin](smi_ssed_130.bin)
|
| 39 |
+
|
| 40 |
+
For more information contact: [email protected] or [email protected].
|
| 41 |
|
| 42 |
## Table of Contents
|
| 43 |
|