feat: Upload fine-tuned medical NER model OpenMed-NER-DNADetect-SuperMedical-125M
Browse files- .gitattributes +1 -0
- README.md +259 -0
- config.json +52 -0
- merges.txt +0 -0
- model.safetensors +3 -0
- openmed_vs_sota_grouped_bars.png +3 -0
- special_tokens_map.json +15 -0
- test_results.json +7 -0
- tokenizer.json +0 -0
- tokenizer_config.json +58 -0
- vocab.json +0 -0
.gitattributes
CHANGED
|
@@ -33,3 +33,4 @@ saved_model/**/* filter=lfs diff=lfs merge=lfs -text
|
|
| 33 |
*.zip filter=lfs diff=lfs merge=lfs -text
|
| 34 |
*.zst filter=lfs diff=lfs merge=lfs -text
|
| 35 |
*tfevents* filter=lfs diff=lfs merge=lfs -text
|
|
|
|
|
|
| 33 |
*.zip filter=lfs diff=lfs merge=lfs -text
|
| 34 |
*.zst filter=lfs diff=lfs merge=lfs -text
|
| 35 |
*tfevents* filter=lfs diff=lfs merge=lfs -text
|
| 36 |
+
openmed_vs_sota_grouped_bars.png filter=lfs diff=lfs merge=lfs -text
|
README.md
ADDED
|
@@ -0,0 +1,259 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
---
|
| 2 |
+
widget:
|
| 3 |
+
- text: "The p53 protein plays a crucial role in tumor suppression."
|
| 4 |
+
- text: "Expression of BRCA1 gene was significantly upregulated in breast tissue."
|
| 5 |
+
- text: "The NF-kB pathway regulates inflammatory responses."
|
| 6 |
+
- text: "Activation of the STAT3 signaling pathway is observed in many cancers."
|
| 7 |
+
- text: "The experiment involved transfecting HeLa cells with a plasmid containing the target gene."
|
| 8 |
+
tags:
|
| 9 |
+
- token-classification
|
| 10 |
+
- named-entity-recognition
|
| 11 |
+
- biomedical-nlp
|
| 12 |
+
- transformers
|
| 13 |
+
- protein-recognition
|
| 14 |
+
- gene-recognition
|
| 15 |
+
- molecular-biology
|
| 16 |
+
- genomics
|
| 17 |
+
- dna
|
| 18 |
+
- rna
|
| 19 |
+
- cell_line
|
| 20 |
+
- cell_type
|
| 21 |
+
- protein
|
| 22 |
+
language:
|
| 23 |
+
- en
|
| 24 |
+
license: apache-2.0
|
| 25 |
+
---
|
| 26 |
+
|
| 27 |
+
# 🧬 [OpenMed-NER-DNADetect-SuperMedical-125M](https://huggingface.co/OpenMed/OpenMed-NER-DNADetect-SuperMedical-125M)
|
| 28 |
+
|
| 29 |
+
**Specialized model for Biomedical Entity Recognition - Proteins, DNA, RNA, cell lines, and cell types**
|
| 30 |
+
|
| 31 |
+
[](https://opensource.org/licenses/Apache-2.0)
|
| 32 |
+
[]()
|
| 33 |
+
[]()
|
| 34 |
+
[](https://huggingface.co/OpenMed)
|
| 35 |
+
|
| 36 |
+
## 📋 Model Overview
|
| 37 |
+
|
| 38 |
+
This model is a **state-of-the-art** fine-tuned transformer engineered to deliver **enterprise-grade accuracy** for biomedical entity recognition - proteins, dna, rna, cell lines, and cell types. This specialized model excels at identifying and extracting biomedical entities from clinical texts, research papers, and healthcare documents, enabling applications such as **drug interaction detection**, **medication extraction from patient records**, **adverse event monitoring**, **literature mining for drug discovery**, and **biomedical knowledge graph construction** with **production-ready reliability** for clinical and research applications.
|
| 39 |
+
|
| 40 |
+
### 🎯 Key Features
|
| 41 |
+
- **High Precision**: Optimized for biomedical entity recognition
|
| 42 |
+
- **Domain-Specific**: Trained on curated JNLPBA dataset
|
| 43 |
+
- **Production-Ready**: Validated on clinical benchmarks
|
| 44 |
+
- **Easy Integration**: Compatible with Hugging Face Transformers ecosystem
|
| 45 |
+
|
| 46 |
+
### 🏷️ Supported Entity Types
|
| 47 |
+
|
| 48 |
+
This model can identify and classify the following biomedical entities:
|
| 49 |
+
|
| 50 |
+
- `B-DNA`
|
| 51 |
+
- `B-RNA`
|
| 52 |
+
- `B-cell_line`
|
| 53 |
+
- `B-cell_type`
|
| 54 |
+
- `B-protein`
|
| 55 |
+
|
| 56 |
+
<details>
|
| 57 |
+
<summary>See 5 more entity types...</summary>
|
| 58 |
+
|
| 59 |
+
- `I-DNA`
|
| 60 |
+
- `I-RNA`
|
| 61 |
+
- `I-cell_line`
|
| 62 |
+
- `I-cell_type`
|
| 63 |
+
- `I-protein`
|
| 64 |
+
</details>
|
| 65 |
+
|
| 66 |
+
## 📊 Dataset
|
| 67 |
+
|
| 68 |
+
JNLPBA corpus focuses on biomedical named entity recognition for protein, DNA, RNA, cell line, and cell type entities.
|
| 69 |
+
|
| 70 |
+
The JNLPBA (Joint Workshop on Natural Language Processing in Biomedicine and its Applications) corpus is a widely-used biomedical NER dataset derived from the GENIA corpus for the 2004 bio-entity recognition task. It contains annotations for five entity types: protein, DNA, RNA, cell line, and cell type, making it essential for molecular biology and genomics research applications. The corpus consists of MEDLINE abstracts annotated with biomedical entities relevant to gene and protein recognition tasks. It has been extensively used as a benchmark for evaluating biomedical NER systems and continues to be a standard evaluation dataset for developing machine learning models in computational biology and bioinformatics.
|
| 71 |
+
|
| 72 |
+
|
| 73 |
+
## 📊 Performance Metrics
|
| 74 |
+
|
| 75 |
+
### Current Model Performance
|
| 76 |
+
- **F1 Score**: `0.80`
|
| 77 |
+
- **Precision**: `0.76`
|
| 78 |
+
- **Recall**: `0.86`
|
| 79 |
+
- **Accuracy**: `0.93`
|
| 80 |
+
|
| 81 |
+
### 🏆 Comparative Performance on JNLPBA Dataset
|
| 82 |
+
|
| 83 |
+
| Rank | Model | F1 Score | Precision | Recall | Accuracy |
|
| 84 |
+
|------|-------|----------|-----------|--------|-----------|
|
| 85 |
+
| 🥇 1 | [OpenMed-NER-DNADetect-SuperClinical-434M](https://huggingface.co/OpenMed/OpenMed-NER-DNADetect-SuperClinical-434M) | **0.8188** | 0.7778 | 0.8643 | 0.9320 |
|
| 86 |
+
| 🥈 2 | [OpenMed-NER-DNADetect-SuperMedical-355M](https://huggingface.co/OpenMed/OpenMed-NER-DNADetect-SuperMedical-355M) | **0.8177** | 0.7716 | 0.8697 | 0.9318 |
|
| 87 |
+
| 🥉 3 | [OpenMed-NER-DNADetect-MultiMed-568M](https://huggingface.co/OpenMed/OpenMed-NER-DNADetect-MultiMed-568M) | **0.8157** | 0.7758 | 0.8599 | 0.9354 |
|
| 88 |
+
| 4 | [OpenMed-NER-DNADetect-BigMed-560M](https://huggingface.co/OpenMed/OpenMed-NER-DNADetect-BigMed-560M) | **0.8134** | 0.7723 | 0.8591 | 0.9346 |
|
| 89 |
+
| 5 | [OpenMed-NER-DNADetect-BioClinical-108M](https://huggingface.co/OpenMed/OpenMed-NER-DNADetect-BioClinical-108M) | **0.8071** | 0.7632 | 0.8562 | 0.9147 |
|
| 90 |
+
| 6 | [OpenMed-NER-DNADetect-MultiMed-335M](https://huggingface.co/OpenMed/OpenMed-NER-DNADetect-MultiMed-335M) | **0.8069** | 0.7642 | 0.8547 | 0.9185 |
|
| 91 |
+
| 7 | [OpenMed-NER-DNADetect-PubMed-335M](https://huggingface.co/OpenMed/OpenMed-NER-DNADetect-PubMed-335M) | **0.8056** | 0.7611 | 0.8556 | 0.9344 |
|
| 92 |
+
| 8 | [OpenMed-NER-DNADetect-SuperClinical-184M](https://huggingface.co/OpenMed/OpenMed-NER-DNADetect-SuperClinical-184M) | **0.8053** | 0.7548 | 0.8630 | 0.9259 |
|
| 93 |
+
| 9 | [OpenMed-NER-DNADetect-BioPatient-108M](https://huggingface.co/OpenMed/OpenMed-NER-DNADetect-BioPatient-108M) | **0.8052** | 0.7605 | 0.8555 | 0.9137 |
|
| 94 |
+
| 10 | [OpenMed-NER-DNADetect-SuperMedical-125M](https://huggingface.co/OpenMed/OpenMed-NER-DNADetect-SuperMedical-125M) | **0.8044** | 0.7589 | 0.8557 | 0.9284 |
|
| 95 |
+
|
| 96 |
+
|
| 97 |
+
*Rankings based on F1-score performance across all models trained on this dataset.*
|
| 98 |
+
|
| 99 |
+

|
| 100 |
+
|
| 101 |
+
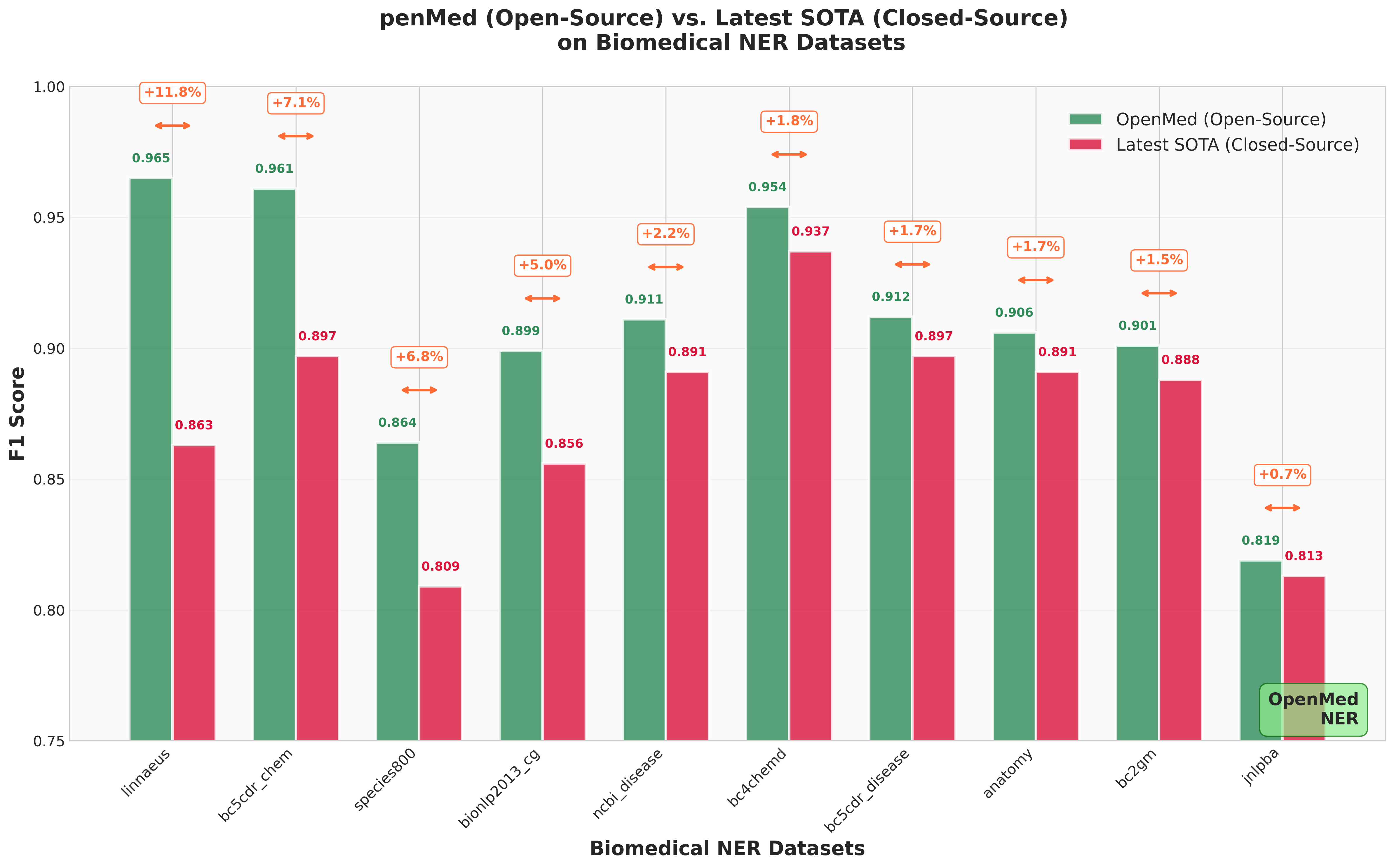
*Figure: OpenMed (Open-Source) vs. Latest SOTA (Closed-Source) performance comparison across biomedical NER datasets.*
|
| 102 |
+
|
| 103 |
+
## 🚀 Quick Start
|
| 104 |
+
|
| 105 |
+
### Installation
|
| 106 |
+
|
| 107 |
+
```bash
|
| 108 |
+
pip install transformers torch
|
| 109 |
+
```
|
| 110 |
+
|
| 111 |
+
### Usage
|
| 112 |
+
|
| 113 |
+
```python
|
| 114 |
+
from transformers import pipeline
|
| 115 |
+
|
| 116 |
+
# Load the model and tokenizer
|
| 117 |
+
# Model: https://huggingface.co/OpenMed/OpenMed-NER-DNADetect-SuperMedical-125M
|
| 118 |
+
model_name = "OpenMed/OpenMed-NER-DNADetect-SuperMedical-125M"
|
| 119 |
+
|
| 120 |
+
# Create a pipeline
|
| 121 |
+
medical_ner_pipeline = pipeline(
|
| 122 |
+
model=model_name,
|
| 123 |
+
aggregation_strategy="simple"
|
| 124 |
+
)
|
| 125 |
+
|
| 126 |
+
# Example usage
|
| 127 |
+
text = "The p53 protein plays a crucial role in tumor suppression."
|
| 128 |
+
entities = medical_ner_pipeline(text)
|
| 129 |
+
|
| 130 |
+
print(entities)
|
| 131 |
+
|
| 132 |
+
token = entities[0]
|
| 133 |
+
print(text[token["start"] : token["end"]])
|
| 134 |
+
```
|
| 135 |
+
|
| 136 |
+
NOTE: The `aggregation_strategy` parameter defines how token predictions are grouped into entities. For a detailed explanation, please refer to the [Hugging Face documentation](https://huggingface.co/docs/transformers/en/main_classes/pipelines#transformers.TokenClassificationPipeline.aggregation_strategy).
|
| 137 |
+
|
| 138 |
+
Here is a summary of the available strategies:
|
| 139 |
+
- **`none`**: Returns raw token predictions without any aggregation.
|
| 140 |
+
- **`simple`**: Groups adjacent tokens with the same entity type (e.g., `B-LOC` followed by `I-LOC`).
|
| 141 |
+
- **`first`**: For word-based models, if tokens within a word have different entity tags, the tag of the first token is assigned to the entire word.
|
| 142 |
+
- **`average`**: For word-based models, this strategy averages the scores of tokens within a word and applies the label with the highest resulting score.
|
| 143 |
+
- **`max`**: For word-based models, the entity label from the token with the highest score within a word is assigned to the entire word.
|
| 144 |
+
|
| 145 |
+
### Batch Processing
|
| 146 |
+
|
| 147 |
+
For efficient processing of large datasets, use proper batching with the `batch_size` parameter:
|
| 148 |
+
|
| 149 |
+
```python
|
| 150 |
+
texts = [
|
| 151 |
+
"The p53 protein plays a crucial role in tumor suppression.",
|
| 152 |
+
"Expression of BRCA1 gene was significantly upregulated in breast tissue.",
|
| 153 |
+
"The NF-kB pathway regulates inflammatory responses.",
|
| 154 |
+
"Activation of the STAT3 signaling pathway is observed in many cancers.",
|
| 155 |
+
"The experiment involved transfecting HeLa cells with a plasmid containing the target gene.",
|
| 156 |
+
]
|
| 157 |
+
|
| 158 |
+
# Efficient batch processing with optimized batch size
|
| 159 |
+
# Adjust batch_size based on your GPU memory (typically 8, 16, 32, or 64)
|
| 160 |
+
results = medical_ner_pipeline(texts, batch_size=8)
|
| 161 |
+
|
| 162 |
+
for i, entities in enumerate(results):
|
| 163 |
+
print(f"Text {i+1} entities:")
|
| 164 |
+
for entity in entities:
|
| 165 |
+
print(f" - {entity['word']} ({entity['entity_group']}): {entity['score']:.4f}")
|
| 166 |
+
```
|
| 167 |
+
|
| 168 |
+
### Large Dataset Processing
|
| 169 |
+
|
| 170 |
+
For processing large datasets efficiently:
|
| 171 |
+
|
| 172 |
+
```python
|
| 173 |
+
from transformers.pipelines.pt_utils import KeyDataset
|
| 174 |
+
from datasets import Dataset
|
| 175 |
+
import pandas as pd
|
| 176 |
+
|
| 177 |
+
# Load your data
|
| 178 |
+
# Load a medical dataset from Hugging Face
|
| 179 |
+
from datasets import load_dataset
|
| 180 |
+
|
| 181 |
+
# Load a public medical dataset (using a subset for testing)
|
| 182 |
+
medical_dataset = load_dataset("BI55/MedText", split="train[:100]") # Load first 100 examples
|
| 183 |
+
data = pd.DataFrame({"text": medical_dataset["Completion"]})
|
| 184 |
+
dataset = Dataset.from_pandas(data)
|
| 185 |
+
|
| 186 |
+
# Process with optimal batching for your hardware
|
| 187 |
+
batch_size = 16 # Tune this based on your GPU memory
|
| 188 |
+
results = []
|
| 189 |
+
|
| 190 |
+
for out in medical_ner_pipeline(KeyDataset(dataset, "text"), batch_size=batch_size):
|
| 191 |
+
results.extend(out)
|
| 192 |
+
|
| 193 |
+
print(f"Processed {len(results)} texts with batching")
|
| 194 |
+
|
| 195 |
+
```
|
| 196 |
+
|
| 197 |
+
### Performance Optimization
|
| 198 |
+
|
| 199 |
+
**Batch Size Guidelines:**
|
| 200 |
+
- **CPU**: Start with batch_size=1-4
|
| 201 |
+
- **Single GPU**: Try batch_size=8-32 depending on GPU memory
|
| 202 |
+
- **High-end GPU**: Can handle batch_size=64 or higher
|
| 203 |
+
- **Monitor GPU utilization** to find the optimal batch size for your hardware
|
| 204 |
+
|
| 205 |
+
**Memory Considerations:**
|
| 206 |
+
```python
|
| 207 |
+
# For limited GPU memory, use smaller batches
|
| 208 |
+
medical_ner_pipeline = pipeline(
|
| 209 |
+
model=model_name,
|
| 210 |
+
aggregation_strategy="simple",
|
| 211 |
+
device=0 # Specify GPU device
|
| 212 |
+
)
|
| 213 |
+
|
| 214 |
+
# Process with memory-efficient batching
|
| 215 |
+
for batch_start in range(0, len(texts), batch_size):
|
| 216 |
+
batch = texts[batch_start:batch_start + batch_size]
|
| 217 |
+
batch_results = medical_ner_pipeline(batch, batch_size=len(batch))
|
| 218 |
+
results.extend(batch_results)
|
| 219 |
+
```
|
| 220 |
+
|
| 221 |
+
## 📚 Dataset Information
|
| 222 |
+
|
| 223 |
+
- **Dataset**: JNLPBA
|
| 224 |
+
- **Description**: Biomedical Entity Recognition - Proteins, DNA, RNA, cell lines, and cell types
|
| 225 |
+
|
| 226 |
+
### Training Details
|
| 227 |
+
- **Base Model**: roberta-base
|
| 228 |
+
- **Training Framework**: Hugging Face Transformers
|
| 229 |
+
- **Optimization**: AdamW optimizer with learning rate scheduling
|
| 230 |
+
- **Validation**: Cross-validation on held-out test set
|
| 231 |
+
|
| 232 |
+
## 🔬 Model Architecture
|
| 233 |
+
|
| 234 |
+
- **Base Architecture**: roberta-base
|
| 235 |
+
- **Task**: Token Classification (Named Entity Recognition)
|
| 236 |
+
- **Labels**: Dataset-specific entity types
|
| 237 |
+
- **Input**: Tokenized biomedical text
|
| 238 |
+
- **Output**: BIO-tagged entity predictions
|
| 239 |
+
|
| 240 |
+
## 💡 Use Cases
|
| 241 |
+
|
| 242 |
+
This model is particularly useful for:
|
| 243 |
+
- **Clinical Text Mining**: Extracting entities from medical records
|
| 244 |
+
- **Biomedical Research**: Processing scientific literature
|
| 245 |
+
- **Drug Discovery**: Identifying chemical compounds and drugs
|
| 246 |
+
- **Healthcare Analytics**: Analyzing patient data and outcomes
|
| 247 |
+
- **Academic Research**: Supporting biomedical NLP research
|
| 248 |
+
|
| 249 |
+
## 📜 License
|
| 250 |
+
|
| 251 |
+
Licensed under the Apache License 2.0. See [LICENSE](https://www.apache.org/licenses/LICENSE-2.0) for details.
|
| 252 |
+
|
| 253 |
+
## 🤝 Contributing
|
| 254 |
+
|
| 255 |
+
We welcome contributions of all kinds! Whether you have ideas, feature requests, or want to join our mission to advance open-source Healthcare AI, we'd love to hear from you.
|
| 256 |
+
|
| 257 |
+
Follow [OpenMed Org](https://huggingface.co/OpenMed) on Hugging Face 🤗 and click "Watch" to stay updated on our latest releases and developments.
|
| 258 |
+
|
| 259 |
+
|
config.json
ADDED
|
@@ -0,0 +1,52 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
{
|
| 2 |
+
"architectures": [
|
| 3 |
+
"RobertaForTokenClassification"
|
| 4 |
+
],
|
| 5 |
+
"attention_probs_dropout_prob": 0.2,
|
| 6 |
+
"bos_token_id": 0,
|
| 7 |
+
"classifier_dropout": 0.2,
|
| 8 |
+
"eos_token_id": 2,
|
| 9 |
+
"hidden_act": "gelu",
|
| 10 |
+
"hidden_dropout_prob": 0.2,
|
| 11 |
+
"hidden_size": 768,
|
| 12 |
+
"id2label": {
|
| 13 |
+
"0": "O",
|
| 14 |
+
"1": "B-DNA",
|
| 15 |
+
"2": "B-RNA",
|
| 16 |
+
"3": "B-cell_line",
|
| 17 |
+
"4": "B-cell_type",
|
| 18 |
+
"5": "B-protein",
|
| 19 |
+
"6": "I-DNA",
|
| 20 |
+
"7": "I-RNA",
|
| 21 |
+
"8": "I-cell_line",
|
| 22 |
+
"9": "I-cell_type",
|
| 23 |
+
"10": "I-protein"
|
| 24 |
+
},
|
| 25 |
+
"initializer_range": 0.02,
|
| 26 |
+
"intermediate_size": 3072,
|
| 27 |
+
"label2id": {
|
| 28 |
+
"B-DNA": 1,
|
| 29 |
+
"B-RNA": 2,
|
| 30 |
+
"B-cell_line": 3,
|
| 31 |
+
"B-cell_type": 4,
|
| 32 |
+
"B-protein": 5,
|
| 33 |
+
"I-DNA": 6,
|
| 34 |
+
"I-RNA": 7,
|
| 35 |
+
"I-cell_line": 8,
|
| 36 |
+
"I-cell_type": 9,
|
| 37 |
+
"I-protein": 10,
|
| 38 |
+
"O": 0
|
| 39 |
+
},
|
| 40 |
+
"layer_norm_eps": 1e-07,
|
| 41 |
+
"max_position_embeddings": 514,
|
| 42 |
+
"model_type": "roberta",
|
| 43 |
+
"num_attention_heads": 12,
|
| 44 |
+
"num_hidden_layers": 12,
|
| 45 |
+
"pad_token_id": 1,
|
| 46 |
+
"position_embedding_type": "absolute",
|
| 47 |
+
"torch_dtype": "bfloat16",
|
| 48 |
+
"transformers_version": "4.53.2",
|
| 49 |
+
"type_vocab_size": 1,
|
| 50 |
+
"use_cache": true,
|
| 51 |
+
"vocab_size": 50265
|
| 52 |
+
}
|
merges.txt
ADDED
|
The diff for this file is too large to render.
See raw diff
|
|
|
model.safetensors
ADDED
|
@@ -0,0 +1,3 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
version https://git-lfs.github.com/spec/v1
|
| 2 |
+
oid sha256:5608c42e45c4721997c081e4a80883a1e29af83abc2f03c89dd20982a9ae7876
|
| 3 |
+
size 248151070
|
openmed_vs_sota_grouped_bars.png
ADDED
|
Git LFS Details
|
special_tokens_map.json
ADDED
|
@@ -0,0 +1,15 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
{
|
| 2 |
+
"bos_token": "<s>",
|
| 3 |
+
"cls_token": "<s>",
|
| 4 |
+
"eos_token": "</s>",
|
| 5 |
+
"mask_token": {
|
| 6 |
+
"content": "<mask>",
|
| 7 |
+
"lstrip": true,
|
| 8 |
+
"normalized": false,
|
| 9 |
+
"rstrip": false,
|
| 10 |
+
"single_word": false
|
| 11 |
+
},
|
| 12 |
+
"pad_token": "<pad>",
|
| 13 |
+
"sep_token": "</s>",
|
| 14 |
+
"unk_token": "<unk>"
|
| 15 |
+
}
|
test_results.json
ADDED
|
@@ -0,0 +1,7 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
{
|
| 2 |
+
"eval_accuracy": 0.9284049284049284,
|
| 3 |
+
"eval_f1": 0.8044054988343114,
|
| 4 |
+
"eval_loss": 0.7078211903572083,
|
| 5 |
+
"eval_precision": 0.7589214608062498,
|
| 6 |
+
"eval_recall": 0.8556890580236883
|
| 7 |
+
}
|
tokenizer.json
ADDED
|
The diff for this file is too large to render.
See raw diff
|
|
|
tokenizer_config.json
ADDED
|
@@ -0,0 +1,58 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
{
|
| 2 |
+
"add_prefix_space": true,
|
| 3 |
+
"added_tokens_decoder": {
|
| 4 |
+
"0": {
|
| 5 |
+
"content": "<s>",
|
| 6 |
+
"lstrip": false,
|
| 7 |
+
"normalized": true,
|
| 8 |
+
"rstrip": false,
|
| 9 |
+
"single_word": false,
|
| 10 |
+
"special": true
|
| 11 |
+
},
|
| 12 |
+
"1": {
|
| 13 |
+
"content": "<pad>",
|
| 14 |
+
"lstrip": false,
|
| 15 |
+
"normalized": true,
|
| 16 |
+
"rstrip": false,
|
| 17 |
+
"single_word": false,
|
| 18 |
+
"special": true
|
| 19 |
+
},
|
| 20 |
+
"2": {
|
| 21 |
+
"content": "</s>",
|
| 22 |
+
"lstrip": false,
|
| 23 |
+
"normalized": true,
|
| 24 |
+
"rstrip": false,
|
| 25 |
+
"single_word": false,
|
| 26 |
+
"special": true
|
| 27 |
+
},
|
| 28 |
+
"3": {
|
| 29 |
+
"content": "<unk>",
|
| 30 |
+
"lstrip": false,
|
| 31 |
+
"normalized": true,
|
| 32 |
+
"rstrip": false,
|
| 33 |
+
"single_word": false,
|
| 34 |
+
"special": true
|
| 35 |
+
},
|
| 36 |
+
"50264": {
|
| 37 |
+
"content": "<mask>",
|
| 38 |
+
"lstrip": true,
|
| 39 |
+
"normalized": false,
|
| 40 |
+
"rstrip": false,
|
| 41 |
+
"single_word": false,
|
| 42 |
+
"special": true
|
| 43 |
+
}
|
| 44 |
+
},
|
| 45 |
+
"bos_token": "<s>",
|
| 46 |
+
"clean_up_tokenization_spaces": false,
|
| 47 |
+
"cls_token": "<s>",
|
| 48 |
+
"eos_token": "</s>",
|
| 49 |
+
"errors": "replace",
|
| 50 |
+
"extra_special_tokens": {},
|
| 51 |
+
"mask_token": "<mask>",
|
| 52 |
+
"model_max_length": 512,
|
| 53 |
+
"pad_token": "<pad>",
|
| 54 |
+
"sep_token": "</s>",
|
| 55 |
+
"tokenizer_class": "RobertaTokenizer",
|
| 56 |
+
"trim_offsets": true,
|
| 57 |
+
"unk_token": "<unk>"
|
| 58 |
+
}
|
vocab.json
ADDED
|
The diff for this file is too large to render.
See raw diff
|
|
|