Upload pathology_nuclei_classification version 0.2.1
Browse files- .gitattributes +1 -0
- LICENSE +201 -0
- configs/evaluate.json +77 -0
- configs/inference.json +120 -0
- configs/inference_trt.json +6 -0
- configs/logging.conf +21 -0
- configs/metadata.json +108 -0
- configs/multi_gpu_evaluate.json +36 -0
- configs/multi_gpu_train.json +44 -0
- configs/train.json +364 -0
- docs/README.md +232 -0
- docs/data_license.txt +6 -0
- models/model.pt +3 -0
- models/model.ts +3 -0
- scripts/__init__.py +13 -0
- scripts/data_process.py +79 -0
- scripts/dataset.py +189 -0
- scripts/handlers.py +141 -0
- scripts/writer.py +54 -0
.gitattributes
CHANGED
|
@@ -33,3 +33,4 @@ saved_model/**/* filter=lfs diff=lfs merge=lfs -text
|
|
| 33 |
*.zip filter=lfs diff=lfs merge=lfs -text
|
| 34 |
*.zst filter=lfs diff=lfs merge=lfs -text
|
| 35 |
*tfevents* filter=lfs diff=lfs merge=lfs -text
|
|
|
|
|
|
| 33 |
*.zip filter=lfs diff=lfs merge=lfs -text
|
| 34 |
*.zst filter=lfs diff=lfs merge=lfs -text
|
| 35 |
*tfevents* filter=lfs diff=lfs merge=lfs -text
|
| 36 |
+
models/model.ts filter=lfs diff=lfs merge=lfs -text
|
LICENSE
ADDED
|
@@ -0,0 +1,201 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
Apache License
|
| 2 |
+
Version 2.0, January 2004
|
| 3 |
+
http://www.apache.org/licenses/
|
| 4 |
+
|
| 5 |
+
TERMS AND CONDITIONS FOR USE, REPRODUCTION, AND DISTRIBUTION
|
| 6 |
+
|
| 7 |
+
1. Definitions.
|
| 8 |
+
|
| 9 |
+
"License" shall mean the terms and conditions for use, reproduction,
|
| 10 |
+
and distribution as defined by Sections 1 through 9 of this document.
|
| 11 |
+
|
| 12 |
+
"Licensor" shall mean the copyright owner or entity authorized by
|
| 13 |
+
the copyright owner that is granting the License.
|
| 14 |
+
|
| 15 |
+
"Legal Entity" shall mean the union of the acting entity and all
|
| 16 |
+
other entities that control, are controlled by, or are under common
|
| 17 |
+
control with that entity. For the purposes of this definition,
|
| 18 |
+
"control" means (i) the power, direct or indirect, to cause the
|
| 19 |
+
direction or management of such entity, whether by contract or
|
| 20 |
+
otherwise, or (ii) ownership of fifty percent (50%) or more of the
|
| 21 |
+
outstanding shares, or (iii) beneficial ownership of such entity.
|
| 22 |
+
|
| 23 |
+
"You" (or "Your") shall mean an individual or Legal Entity
|
| 24 |
+
exercising permissions granted by this License.
|
| 25 |
+
|
| 26 |
+
"Source" form shall mean the preferred form for making modifications,
|
| 27 |
+
including but not limited to software source code, documentation
|
| 28 |
+
source, and configuration files.
|
| 29 |
+
|
| 30 |
+
"Object" form shall mean any form resulting from mechanical
|
| 31 |
+
transformation or translation of a Source form, including but
|
| 32 |
+
not limited to compiled object code, generated documentation,
|
| 33 |
+
and conversions to other media types.
|
| 34 |
+
|
| 35 |
+
"Work" shall mean the work of authorship, whether in Source or
|
| 36 |
+
Object form, made available under the License, as indicated by a
|
| 37 |
+
copyright notice that is included in or attached to the work
|
| 38 |
+
(an example is provided in the Appendix below).
|
| 39 |
+
|
| 40 |
+
"Derivative Works" shall mean any work, whether in Source or Object
|
| 41 |
+
form, that is based on (or derived from) the Work and for which the
|
| 42 |
+
editorial revisions, annotations, elaborations, or other modifications
|
| 43 |
+
represent, as a whole, an original work of authorship. For the purposes
|
| 44 |
+
of this License, Derivative Works shall not include works that remain
|
| 45 |
+
separable from, or merely link (or bind by name) to the interfaces of,
|
| 46 |
+
the Work and Derivative Works thereof.
|
| 47 |
+
|
| 48 |
+
"Contribution" shall mean any work of authorship, including
|
| 49 |
+
the original version of the Work and any modifications or additions
|
| 50 |
+
to that Work or Derivative Works thereof, that is intentionally
|
| 51 |
+
submitted to Licensor for inclusion in the Work by the copyright owner
|
| 52 |
+
or by an individual or Legal Entity authorized to submit on behalf of
|
| 53 |
+
the copyright owner. For the purposes of this definition, "submitted"
|
| 54 |
+
means any form of electronic, verbal, or written communication sent
|
| 55 |
+
to the Licensor or its representatives, including but not limited to
|
| 56 |
+
communication on electronic mailing lists, source code control systems,
|
| 57 |
+
and issue tracking systems that are managed by, or on behalf of, the
|
| 58 |
+
Licensor for the purpose of discussing and improving the Work, but
|
| 59 |
+
excluding communication that is conspicuously marked or otherwise
|
| 60 |
+
designated in writing by the copyright owner as "Not a Contribution."
|
| 61 |
+
|
| 62 |
+
"Contributor" shall mean Licensor and any individual or Legal Entity
|
| 63 |
+
on behalf of whom a Contribution has been received by Licensor and
|
| 64 |
+
subsequently incorporated within the Work.
|
| 65 |
+
|
| 66 |
+
2. Grant of Copyright License. Subject to the terms and conditions of
|
| 67 |
+
this License, each Contributor hereby grants to You a perpetual,
|
| 68 |
+
worldwide, non-exclusive, no-charge, royalty-free, irrevocable
|
| 69 |
+
copyright license to reproduce, prepare Derivative Works of,
|
| 70 |
+
publicly display, publicly perform, sublicense, and distribute the
|
| 71 |
+
Work and such Derivative Works in Source or Object form.
|
| 72 |
+
|
| 73 |
+
3. Grant of Patent License. Subject to the terms and conditions of
|
| 74 |
+
this License, each Contributor hereby grants to You a perpetual,
|
| 75 |
+
worldwide, non-exclusive, no-charge, royalty-free, irrevocable
|
| 76 |
+
(except as stated in this section) patent license to make, have made,
|
| 77 |
+
use, offer to sell, sell, import, and otherwise transfer the Work,
|
| 78 |
+
where such license applies only to those patent claims licensable
|
| 79 |
+
by such Contributor that are necessarily infringed by their
|
| 80 |
+
Contribution(s) alone or by combination of their Contribution(s)
|
| 81 |
+
with the Work to which such Contribution(s) was submitted. If You
|
| 82 |
+
institute patent litigation against any entity (including a
|
| 83 |
+
cross-claim or counterclaim in a lawsuit) alleging that the Work
|
| 84 |
+
or a Contribution incorporated within the Work constitutes direct
|
| 85 |
+
or contributory patent infringement, then any patent licenses
|
| 86 |
+
granted to You under this License for that Work shall terminate
|
| 87 |
+
as of the date such litigation is filed.
|
| 88 |
+
|
| 89 |
+
4. Redistribution. You may reproduce and distribute copies of the
|
| 90 |
+
Work or Derivative Works thereof in any medium, with or without
|
| 91 |
+
modifications, and in Source or Object form, provided that You
|
| 92 |
+
meet the following conditions:
|
| 93 |
+
|
| 94 |
+
(a) You must give any other recipients of the Work or
|
| 95 |
+
Derivative Works a copy of this License; and
|
| 96 |
+
|
| 97 |
+
(b) You must cause any modified files to carry prominent notices
|
| 98 |
+
stating that You changed the files; and
|
| 99 |
+
|
| 100 |
+
(c) You must retain, in the Source form of any Derivative Works
|
| 101 |
+
that You distribute, all copyright, patent, trademark, and
|
| 102 |
+
attribution notices from the Source form of the Work,
|
| 103 |
+
excluding those notices that do not pertain to any part of
|
| 104 |
+
the Derivative Works; and
|
| 105 |
+
|
| 106 |
+
(d) If the Work includes a "NOTICE" text file as part of its
|
| 107 |
+
distribution, then any Derivative Works that You distribute must
|
| 108 |
+
include a readable copy of the attribution notices contained
|
| 109 |
+
within such NOTICE file, excluding those notices that do not
|
| 110 |
+
pertain to any part of the Derivative Works, in at least one
|
| 111 |
+
of the following places: within a NOTICE text file distributed
|
| 112 |
+
as part of the Derivative Works; within the Source form or
|
| 113 |
+
documentation, if provided along with the Derivative Works; or,
|
| 114 |
+
within a display generated by the Derivative Works, if and
|
| 115 |
+
wherever such third-party notices normally appear. The contents
|
| 116 |
+
of the NOTICE file are for informational purposes only and
|
| 117 |
+
do not modify the License. You may add Your own attribution
|
| 118 |
+
notices within Derivative Works that You distribute, alongside
|
| 119 |
+
or as an addendum to the NOTICE text from the Work, provided
|
| 120 |
+
that such additional attribution notices cannot be construed
|
| 121 |
+
as modifying the License.
|
| 122 |
+
|
| 123 |
+
You may add Your own copyright statement to Your modifications and
|
| 124 |
+
may provide additional or different license terms and conditions
|
| 125 |
+
for use, reproduction, or distribution of Your modifications, or
|
| 126 |
+
for any such Derivative Works as a whole, provided Your use,
|
| 127 |
+
reproduction, and distribution of the Work otherwise complies with
|
| 128 |
+
the conditions stated in this License.
|
| 129 |
+
|
| 130 |
+
5. Submission of Contributions. Unless You explicitly state otherwise,
|
| 131 |
+
any Contribution intentionally submitted for inclusion in the Work
|
| 132 |
+
by You to the Licensor shall be under the terms and conditions of
|
| 133 |
+
this License, without any additional terms or conditions.
|
| 134 |
+
Notwithstanding the above, nothing herein shall supersede or modify
|
| 135 |
+
the terms of any separate license agreement you may have executed
|
| 136 |
+
with Licensor regarding such Contributions.
|
| 137 |
+
|
| 138 |
+
6. Trademarks. This License does not grant permission to use the trade
|
| 139 |
+
names, trademarks, service marks, or product names of the Licensor,
|
| 140 |
+
except as required for reasonable and customary use in describing the
|
| 141 |
+
origin of the Work and reproducing the content of the NOTICE file.
|
| 142 |
+
|
| 143 |
+
7. Disclaimer of Warranty. Unless required by applicable law or
|
| 144 |
+
agreed to in writing, Licensor provides the Work (and each
|
| 145 |
+
Contributor provides its Contributions) on an "AS IS" BASIS,
|
| 146 |
+
WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or
|
| 147 |
+
implied, including, without limitation, any warranties or conditions
|
| 148 |
+
of TITLE, NON-INFRINGEMENT, MERCHANTABILITY, or FITNESS FOR A
|
| 149 |
+
PARTICULAR PURPOSE. You are solely responsible for determining the
|
| 150 |
+
appropriateness of using or redistributing the Work and assume any
|
| 151 |
+
risks associated with Your exercise of permissions under this License.
|
| 152 |
+
|
| 153 |
+
8. Limitation of Liability. In no event and under no legal theory,
|
| 154 |
+
whether in tort (including negligence), contract, or otherwise,
|
| 155 |
+
unless required by applicable law (such as deliberate and grossly
|
| 156 |
+
negligent acts) or agreed to in writing, shall any Contributor be
|
| 157 |
+
liable to You for damages, including any direct, indirect, special,
|
| 158 |
+
incidental, or consequential damages of any character arising as a
|
| 159 |
+
result of this License or out of the use or inability to use the
|
| 160 |
+
Work (including but not limited to damages for loss of goodwill,
|
| 161 |
+
work stoppage, computer failure or malfunction, or any and all
|
| 162 |
+
other commercial damages or losses), even if such Contributor
|
| 163 |
+
has been advised of the possibility of such damages.
|
| 164 |
+
|
| 165 |
+
9. Accepting Warranty or Additional Liability. While redistributing
|
| 166 |
+
the Work or Derivative Works thereof, You may choose to offer,
|
| 167 |
+
and charge a fee for, acceptance of support, warranty, indemnity,
|
| 168 |
+
or other liability obligations and/or rights consistent with this
|
| 169 |
+
License. However, in accepting such obligations, You may act only
|
| 170 |
+
on Your own behalf and on Your sole responsibility, not on behalf
|
| 171 |
+
of any other Contributor, and only if You agree to indemnify,
|
| 172 |
+
defend, and hold each Contributor harmless for any liability
|
| 173 |
+
incurred by, or claims asserted against, such Contributor by reason
|
| 174 |
+
of your accepting any such warranty or additional liability.
|
| 175 |
+
|
| 176 |
+
END OF TERMS AND CONDITIONS
|
| 177 |
+
|
| 178 |
+
APPENDIX: How to apply the Apache License to your work.
|
| 179 |
+
|
| 180 |
+
To apply the Apache License to your work, attach the following
|
| 181 |
+
boilerplate notice, with the fields enclosed by brackets "[]"
|
| 182 |
+
replaced with your own identifying information. (Don't include
|
| 183 |
+
the brackets!) The text should be enclosed in the appropriate
|
| 184 |
+
comment syntax for the file format. We also recommend that a
|
| 185 |
+
file or class name and description of purpose be included on the
|
| 186 |
+
same "printed page" as the copyright notice for easier
|
| 187 |
+
identification within third-party archives.
|
| 188 |
+
|
| 189 |
+
Copyright [yyyy] [name of copyright owner]
|
| 190 |
+
|
| 191 |
+
Licensed under the Apache License, Version 2.0 (the "License");
|
| 192 |
+
you may not use this file except in compliance with the License.
|
| 193 |
+
You may obtain a copy of the License at
|
| 194 |
+
|
| 195 |
+
http://www.apache.org/licenses/LICENSE-2.0
|
| 196 |
+
|
| 197 |
+
Unless required by applicable law or agreed to in writing, software
|
| 198 |
+
distributed under the License is distributed on an "AS IS" BASIS,
|
| 199 |
+
WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.
|
| 200 |
+
See the License for the specific language governing permissions and
|
| 201 |
+
limitations under the License.
|
configs/evaluate.json
ADDED
|
@@ -0,0 +1,77 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
{
|
| 2 |
+
"validate#dataset#cache_rate": 0,
|
| 3 |
+
"validate#postprocessing": {
|
| 4 |
+
"_target_": "Compose",
|
| 5 |
+
"transforms": [
|
| 6 |
+
{
|
| 7 |
+
"_target_": "Activationsd",
|
| 8 |
+
"keys": "pred",
|
| 9 |
+
"softmax": true
|
| 10 |
+
},
|
| 11 |
+
{
|
| 12 |
+
"_target_": "AsDiscreted",
|
| 13 |
+
"keys": [
|
| 14 |
+
"pred",
|
| 15 |
+
"label"
|
| 16 |
+
],
|
| 17 |
+
"argmax": [
|
| 18 |
+
true,
|
| 19 |
+
false
|
| 20 |
+
],
|
| 21 |
+
"to_onehot": 4
|
| 22 |
+
},
|
| 23 |
+
{
|
| 24 |
+
"_target_": "ToTensord",
|
| 25 |
+
"keys": [
|
| 26 |
+
"pred",
|
| 27 |
+
"label"
|
| 28 |
+
],
|
| 29 |
+
"device": "@device"
|
| 30 |
+
},
|
| 31 |
+
{
|
| 32 |
+
"_target_": "SaveImaged",
|
| 33 |
+
"_disabled_": true,
|
| 34 |
+
"keys": "pred",
|
| 35 |
+
"meta_keys": "pred_meta_dict",
|
| 36 |
+
"output_dir": "@output_dir",
|
| 37 |
+
"output_ext": ".json"
|
| 38 |
+
}
|
| 39 |
+
]
|
| 40 |
+
},
|
| 41 |
+
"validate#handlers": [
|
| 42 |
+
{
|
| 43 |
+
"_target_": "CheckpointLoader",
|
| 44 |
+
"load_path": "$@ckpt_dir + '/model.pt'",
|
| 45 |
+
"load_dict": {
|
| 46 |
+
"model": "@network"
|
| 47 |
+
}
|
| 48 |
+
},
|
| 49 |
+
{
|
| 50 |
+
"_target_": "StatsHandler",
|
| 51 |
+
"iteration_log": false
|
| 52 |
+
},
|
| 53 |
+
{
|
| 54 |
+
"_target_": "MetricsSaver",
|
| 55 |
+
"save_dir": "@output_dir",
|
| 56 |
+
"metrics": [
|
| 57 |
+
"val_f1",
|
| 58 |
+
"val_accuracy"
|
| 59 |
+
],
|
| 60 |
+
"metric_details": [
|
| 61 |
+
"val_f1"
|
| 62 |
+
],
|
| 63 |
+
"batch_transform": "$monai.handlers.from_engine(['image_meta_dict'])",
|
| 64 |
+
"summary_ops": "*"
|
| 65 |
+
}
|
| 66 |
+
],
|
| 67 |
+
"initialize": [
|
| 68 |
+
"$import sys",
|
| 69 |
+
"$sys.path.append(@bundle_root)",
|
| 70 |
+
"$monai.utils.set_determinism(seed=123)",
|
| 71 |
+
"$import scripts",
|
| 72 |
+
"$monai.data.register_writer('json', scripts.ClassificationWriter)"
|
| 73 |
+
],
|
| 74 |
+
"run": [
|
| 75 |
+
"$@validate#evaluator.run()"
|
| 76 |
+
]
|
| 77 |
+
}
|
configs/inference.json
ADDED
|
@@ -0,0 +1,120 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
{
|
| 2 |
+
"imports": [
|
| 3 |
+
"$import glob",
|
| 4 |
+
"$import json",
|
| 5 |
+
"$import pathlib",
|
| 6 |
+
"$import os"
|
| 7 |
+
],
|
| 8 |
+
"bundle_root": ".",
|
| 9 |
+
"checkpoint": "$@bundle_root + '/models/model.pt'",
|
| 10 |
+
"output_dir": "$@bundle_root + '/eval'",
|
| 11 |
+
"dataset_dir": "/workspace/data/CoNSePNuclei",
|
| 12 |
+
"images": "$list(sorted(glob.glob(@dataset_dir + '/Test/Images/*.png')))[:1]",
|
| 13 |
+
"labels": "$list(sorted(glob.glob(@dataset_dir + '/Test/Labels/*.png')))[:1]",
|
| 14 |
+
"input_data": "$[{'image': i, 'label': l} for i,l in zip(@images, @labels)]",
|
| 15 |
+
"device": "$torch.device('cuda:0' if torch.cuda.is_available() else 'cpu')",
|
| 16 |
+
"network_def": {
|
| 17 |
+
"_target_": "DenseNet121",
|
| 18 |
+
"spatial_dims": 2,
|
| 19 |
+
"in_channels": 4,
|
| 20 |
+
"out_channels": 4
|
| 21 |
+
},
|
| 22 |
+
"network": "$@network_def.to(@device)",
|
| 23 |
+
"preprocessing": {
|
| 24 |
+
"_target_": "Compose",
|
| 25 |
+
"transforms": [
|
| 26 |
+
{
|
| 27 |
+
"_target_": "LoadImaged",
|
| 28 |
+
"keys": [
|
| 29 |
+
"image",
|
| 30 |
+
"label"
|
| 31 |
+
],
|
| 32 |
+
"dtype": "uint8",
|
| 33 |
+
"image_only": false
|
| 34 |
+
},
|
| 35 |
+
{
|
| 36 |
+
"_target_": "EnsureChannelFirstd",
|
| 37 |
+
"keys": [
|
| 38 |
+
"image",
|
| 39 |
+
"label"
|
| 40 |
+
]
|
| 41 |
+
},
|
| 42 |
+
{
|
| 43 |
+
"_target_": "ScaleIntensityRanged",
|
| 44 |
+
"keys": "image",
|
| 45 |
+
"a_min": 0.0,
|
| 46 |
+
"a_max": 255.0,
|
| 47 |
+
"b_min": -1.0,
|
| 48 |
+
"b_max": 1.0
|
| 49 |
+
},
|
| 50 |
+
{
|
| 51 |
+
"_target_": "AddLabelAsGuidanced",
|
| 52 |
+
"keys": "image",
|
| 53 |
+
"source": "label"
|
| 54 |
+
}
|
| 55 |
+
]
|
| 56 |
+
},
|
| 57 |
+
"dataset": {
|
| 58 |
+
"_target_": "Dataset",
|
| 59 |
+
"data": "@input_data",
|
| 60 |
+
"transform": "@preprocessing"
|
| 61 |
+
},
|
| 62 |
+
"dataloader": {
|
| 63 |
+
"_target_": "DataLoader",
|
| 64 |
+
"dataset": "@dataset",
|
| 65 |
+
"batch_size": 1,
|
| 66 |
+
"shuffle": false,
|
| 67 |
+
"num_workers": 4
|
| 68 |
+
},
|
| 69 |
+
"inferer": {
|
| 70 |
+
"_target_": "SimpleInferer"
|
| 71 |
+
},
|
| 72 |
+
"postprocessing": {
|
| 73 |
+
"_target_": "Compose",
|
| 74 |
+
"transforms": [
|
| 75 |
+
{
|
| 76 |
+
"_target_": "Activationsd",
|
| 77 |
+
"keys": "pred",
|
| 78 |
+
"softmax": true
|
| 79 |
+
},
|
| 80 |
+
{
|
| 81 |
+
"_target_": "SaveImaged",
|
| 82 |
+
"keys": "pred",
|
| 83 |
+
"meta_keys": "pred_meta_dict",
|
| 84 |
+
"output_dir": "@output_dir",
|
| 85 |
+
"output_ext": ".json"
|
| 86 |
+
}
|
| 87 |
+
]
|
| 88 |
+
},
|
| 89 |
+
"handlers": [
|
| 90 |
+
{
|
| 91 |
+
"_target_": "CheckpointLoader",
|
| 92 |
+
"load_path": "$@checkpoint",
|
| 93 |
+
"load_dict": {
|
| 94 |
+
"model": "@network"
|
| 95 |
+
}
|
| 96 |
+
},
|
| 97 |
+
{
|
| 98 |
+
"_target_": "StatsHandler",
|
| 99 |
+
"iteration_log": false
|
| 100 |
+
}
|
| 101 |
+
],
|
| 102 |
+
"evaluator": {
|
| 103 |
+
"_target_": "SupervisedEvaluator",
|
| 104 |
+
"device": "@device",
|
| 105 |
+
"val_data_loader": "@dataloader",
|
| 106 |
+
"network": "@network",
|
| 107 |
+
"inferer": "@inferer",
|
| 108 |
+
"postprocessing": "@postprocessing",
|
| 109 |
+
"val_handlers": "@handlers",
|
| 110 |
+
"amp": true
|
| 111 |
+
},
|
| 112 |
+
"initialize": [
|
| 113 |
+
"$monai.utils.set_determinism(seed=123)",
|
| 114 |
+
"$import scripts",
|
| 115 |
+
"$monai.data.register_writer('json', scripts.ClassificationWriter)"
|
| 116 |
+
],
|
| 117 |
+
"run": [
|
| 118 |
+
"[email protected]()"
|
| 119 |
+
]
|
| 120 |
+
}
|
configs/inference_trt.json
ADDED
|
@@ -0,0 +1,6 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
{
|
| 2 |
+
"+imports": [
|
| 3 |
+
"$from monai.networks import trt_compile"
|
| 4 |
+
],
|
| 5 |
+
"network": "$trt_compile(@network_def.to(@device), @checkpoint)"
|
| 6 |
+
}
|
configs/logging.conf
ADDED
|
@@ -0,0 +1,21 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
[loggers]
|
| 2 |
+
keys=root
|
| 3 |
+
|
| 4 |
+
[handlers]
|
| 5 |
+
keys=consoleHandler
|
| 6 |
+
|
| 7 |
+
[formatters]
|
| 8 |
+
keys=fullFormatter
|
| 9 |
+
|
| 10 |
+
[logger_root]
|
| 11 |
+
level=INFO
|
| 12 |
+
handlers=consoleHandler
|
| 13 |
+
|
| 14 |
+
[handler_consoleHandler]
|
| 15 |
+
class=StreamHandler
|
| 16 |
+
level=INFO
|
| 17 |
+
formatter=fullFormatter
|
| 18 |
+
args=(sys.stdout,)
|
| 19 |
+
|
| 20 |
+
[formatter_fullFormatter]
|
| 21 |
+
format=%(asctime)s - %(name)s - %(levelname)s - %(message)s
|
configs/metadata.json
ADDED
|
@@ -0,0 +1,108 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
{
|
| 2 |
+
"schema": "https://github.com/Project-MONAI/MONAI-extra-test-data/releases/download/0.8.1/meta_schema_20240725.json",
|
| 3 |
+
"version": "0.2.1",
|
| 4 |
+
"changelog": {
|
| 5 |
+
"0.2.1": "update to huggingface hosting",
|
| 6 |
+
"0.2.0": "update issue for IgniteInfo",
|
| 7 |
+
"0.1.9": "update tensorrt benchmark results",
|
| 8 |
+
"0.1.8": "enable tensorrt",
|
| 9 |
+
"0.1.7": "update to use monai 1.3.1",
|
| 10 |
+
"0.1.6": "set image_only to False",
|
| 11 |
+
"0.1.5": "add support for TensorRT conversion and inference",
|
| 12 |
+
"0.1.4": "fix the wrong GPU index issue of multi-node",
|
| 13 |
+
"0.1.3": "remove error dollar symbol in readme",
|
| 14 |
+
"0.1.2": "add RAM warning",
|
| 15 |
+
"0.1.1": "enable deterministic eval and inference",
|
| 16 |
+
"0.1.0": "Update deterministic results",
|
| 17 |
+
"0.0.9": "Update README Formatting",
|
| 18 |
+
"0.0.8": "enable deterministic training",
|
| 19 |
+
"0.0.7": "update benchmark on A100",
|
| 20 |
+
"0.0.6": "adapt to BundleWorkflow interface",
|
| 21 |
+
"0.0.5": "add name tag",
|
| 22 |
+
"0.0.4": "Fix evaluation",
|
| 23 |
+
"0.0.3": "Update to use MONAI 1.1.0",
|
| 24 |
+
"0.0.2": "Update The Torch Vision Transform",
|
| 25 |
+
"0.0.1": "initialize the model package structure"
|
| 26 |
+
},
|
| 27 |
+
"monai_version": "1.4.0",
|
| 28 |
+
"pytorch_version": "2.4.0",
|
| 29 |
+
"numpy_version": "1.24.4",
|
| 30 |
+
"required_packages_version": {
|
| 31 |
+
"nibabel": "5.2.1",
|
| 32 |
+
"pytorch-ignite": "0.4.11",
|
| 33 |
+
"torchvision": "0.19.0",
|
| 34 |
+
"scipy": "1.13.1",
|
| 35 |
+
"scikit-learn": "1.5.1",
|
| 36 |
+
"tensorboard": "2.17.0",
|
| 37 |
+
"scikit-image": "0.23.2"
|
| 38 |
+
},
|
| 39 |
+
"supported_apps": {},
|
| 40 |
+
"name": "Pathology nuclei classification",
|
| 41 |
+
"task": "Pathology Nuclei classification",
|
| 42 |
+
"description": "A pre-trained model for Nuclei Classification within Haematoxylin & Eosin stained histology images",
|
| 43 |
+
"authors": "MONAI team",
|
| 44 |
+
"copyright": "Copyright (c) MONAI Consortium",
|
| 45 |
+
"data_source": "consep_dataset.zip from https://warwick.ac.uk/fac/cross_fac/tia/data/hovernet",
|
| 46 |
+
"data_type": "png",
|
| 47 |
+
"image_classes": "RGB channel data, intensity scaled to [0, 1]",
|
| 48 |
+
"label_classes": "single channel data",
|
| 49 |
+
"pred_classes": "4 channels OneHot data, channel 0 is Other, channel 1 is Inflammatory, channel 2 is Epithelial, channel 3 is Spindle-Shaped",
|
| 50 |
+
"eval_metrics": {
|
| 51 |
+
"f1_score": 0.852
|
| 52 |
+
},
|
| 53 |
+
"intended_use": "This is an example, not to be used for diagnostic purposes",
|
| 54 |
+
"references": [
|
| 55 |
+
"S. Graham, Q. D. Vu, S. E. A. Raza, A. Azam, Y-W. Tsang, J. T. Kwak and N. Rajpoot. \"HoVer-Net: Simultaneous Segmentation and Classification of Nuclei in Multi-Tissue Histology Images.\" Medical Image Analysis, Sept. 2019. https://doi.org/10.1016/j.media.2019.101563"
|
| 56 |
+
],
|
| 57 |
+
"network_data_format": {
|
| 58 |
+
"inputs": {
|
| 59 |
+
"image": {
|
| 60 |
+
"type": "magnitude",
|
| 61 |
+
"format": "RGB",
|
| 62 |
+
"modality": "regular",
|
| 63 |
+
"num_channels": 4,
|
| 64 |
+
"spatial_shape": [
|
| 65 |
+
128,
|
| 66 |
+
128
|
| 67 |
+
],
|
| 68 |
+
"dtype": "float32",
|
| 69 |
+
"value_range": [
|
| 70 |
+
0,
|
| 71 |
+
1
|
| 72 |
+
],
|
| 73 |
+
"is_patch_data": false,
|
| 74 |
+
"channel_def": {
|
| 75 |
+
"0": "R",
|
| 76 |
+
"1": "G",
|
| 77 |
+
"2": "B",
|
| 78 |
+
"3": "Mask"
|
| 79 |
+
}
|
| 80 |
+
}
|
| 81 |
+
},
|
| 82 |
+
"outputs": {
|
| 83 |
+
"pred": {
|
| 84 |
+
"type": "probabilities",
|
| 85 |
+
"format": "classes",
|
| 86 |
+
"num_channels": 4,
|
| 87 |
+
"spatial_shape": [
|
| 88 |
+
1,
|
| 89 |
+
4
|
| 90 |
+
],
|
| 91 |
+
"dtype": "float32",
|
| 92 |
+
"value_range": [
|
| 93 |
+
0,
|
| 94 |
+
1,
|
| 95 |
+
2,
|
| 96 |
+
3
|
| 97 |
+
],
|
| 98 |
+
"is_patch_data": false,
|
| 99 |
+
"channel_def": {
|
| 100 |
+
"0": "Other",
|
| 101 |
+
"1": "Inflammatory",
|
| 102 |
+
"2": "Epithelial",
|
| 103 |
+
"3": "Spindle-Shaped"
|
| 104 |
+
}
|
| 105 |
+
}
|
| 106 |
+
}
|
| 107 |
+
}
|
| 108 |
+
}
|
configs/multi_gpu_evaluate.json
ADDED
|
@@ -0,0 +1,36 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
{
|
| 2 |
+
"device": "$torch.device('cuda:' + os.environ['LOCAL_RANK'])",
|
| 3 |
+
"network": {
|
| 4 |
+
"_target_": "torch.nn.parallel.DistributedDataParallel",
|
| 5 |
+
"module": "$@network_def.to(@device)",
|
| 6 |
+
"device_ids": [
|
| 7 |
+
"@device"
|
| 8 |
+
]
|
| 9 |
+
},
|
| 10 |
+
"validate#sampler": {
|
| 11 |
+
"_target_": "DistributedSampler",
|
| 12 |
+
"dataset": "@validate#dataset",
|
| 13 |
+
"even_divisible": false,
|
| 14 |
+
"shuffle": false
|
| 15 |
+
},
|
| 16 |
+
"validate#dataloader#sampler": "@validate#sampler",
|
| 17 |
+
"validate#handlers#1#_disabled_": "$dist.get_rank() > 0",
|
| 18 |
+
"initialize": [
|
| 19 |
+
"$import sys",
|
| 20 |
+
"$sys.path.append(@bundle_root)",
|
| 21 |
+
"$import torch.distributed as dist",
|
| 22 |
+
"$dist.is_initialized() or dist.init_process_group(backend='nccl')",
|
| 23 |
+
"$torch.cuda.set_device(@device)",
|
| 24 |
+
"$monai.utils.set_determinism(seed=123)",
|
| 25 |
+
"$import logging",
|
| 26 |
+
"$@validate#evaluator.logger.setLevel(logging.WARNING if dist.get_rank() > 0 else logging.INFO)",
|
| 27 |
+
"$import scripts",
|
| 28 |
+
"$monai.data.register_writer('json', scripts.ClassificationWriter)"
|
| 29 |
+
],
|
| 30 |
+
"run": [
|
| 31 |
+
"$@validate#evaluator.run()"
|
| 32 |
+
],
|
| 33 |
+
"finalize": [
|
| 34 |
+
"$dist.is_initialized() and dist.destroy_process_group()"
|
| 35 |
+
]
|
| 36 |
+
}
|
configs/multi_gpu_train.json
ADDED
|
@@ -0,0 +1,44 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
{
|
| 2 |
+
"device": "$torch.device('cuda:' + os.environ['LOCAL_RANK'])",
|
| 3 |
+
"network": {
|
| 4 |
+
"_target_": "torch.nn.parallel.DistributedDataParallel",
|
| 5 |
+
"module": "$@network_def.to(@device)",
|
| 6 |
+
"device_ids": [
|
| 7 |
+
"@device"
|
| 8 |
+
]
|
| 9 |
+
},
|
| 10 |
+
"train#sampler": {
|
| 11 |
+
"_target_": "DistributedSampler",
|
| 12 |
+
"dataset": "@train#dataset",
|
| 13 |
+
"even_divisible": true,
|
| 14 |
+
"shuffle": true
|
| 15 |
+
},
|
| 16 |
+
"train#dataloader#sampler": "@train#sampler",
|
| 17 |
+
"train#dataloader#shuffle": false,
|
| 18 |
+
"train#trainer#train_handlers": "$@train#handlers[: -2 if dist.get_rank() > 0 else None]",
|
| 19 |
+
"validate#sampler": {
|
| 20 |
+
"_target_": "DistributedSampler",
|
| 21 |
+
"dataset": "@validate#dataset",
|
| 22 |
+
"even_divisible": false,
|
| 23 |
+
"shuffle": false
|
| 24 |
+
},
|
| 25 |
+
"validate#dataloader#sampler": "@validate#sampler",
|
| 26 |
+
"validate#evaluator#val_handlers": "$None if dist.get_rank() > 0 else @validate#handlers",
|
| 27 |
+
"initialize": [
|
| 28 |
+
"$import sys",
|
| 29 |
+
"$sys.path.append(@bundle_root)",
|
| 30 |
+
"$import torch.distributed as dist",
|
| 31 |
+
"$dist.is_initialized() or dist.init_process_group(backend='nccl')",
|
| 32 |
+
"$torch.cuda.set_device(@device)",
|
| 33 |
+
"$monai.utils.set_determinism(seed=123)",
|
| 34 |
+
"$import logging",
|
| 35 |
+
"$@train#trainer.logger.setLevel(logging.WARNING if dist.get_rank() > 0 else logging.INFO)",
|
| 36 |
+
"$@validate#evaluator.logger.setLevel(logging.WARNING if dist.get_rank() > 0 else logging.INFO)"
|
| 37 |
+
],
|
| 38 |
+
"run": [
|
| 39 |
+
"$@train#trainer.run()"
|
| 40 |
+
],
|
| 41 |
+
"finalize": [
|
| 42 |
+
"$dist.is_initialized() and dist.destroy_process_group()"
|
| 43 |
+
]
|
| 44 |
+
}
|
configs/train.json
ADDED
|
@@ -0,0 +1,364 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
{
|
| 2 |
+
"imports": [
|
| 3 |
+
"$import glob",
|
| 4 |
+
"$import ignite",
|
| 5 |
+
"$import json",
|
| 6 |
+
"$import pathlib",
|
| 7 |
+
"$import os"
|
| 8 |
+
],
|
| 9 |
+
"bundle_root": ".",
|
| 10 |
+
"ckpt_dir": "$@bundle_root + '/models'",
|
| 11 |
+
"output_dir": "$@bundle_root + '/eval'",
|
| 12 |
+
"dataset_dir": "/workspace/data/CoNSePNuclei",
|
| 13 |
+
"dataset_json": "$@dataset_dir + '/dataset.json'",
|
| 14 |
+
"train_datalist": "$json.loads(pathlib.Path(@dataset_json).read_text())['training']",
|
| 15 |
+
"val_datalist": "$json.loads(pathlib.Path(@dataset_json).read_text())['validation']",
|
| 16 |
+
"val_interval": 1,
|
| 17 |
+
"device": "$torch.device('cuda:0' if torch.cuda.is_available() else 'cpu')",
|
| 18 |
+
"network_def": {
|
| 19 |
+
"_target_": "DenseNet121",
|
| 20 |
+
"spatial_dims": 2,
|
| 21 |
+
"in_channels": 4,
|
| 22 |
+
"out_channels": 4
|
| 23 |
+
},
|
| 24 |
+
"network": "$@network_def.to(@device)",
|
| 25 |
+
"loss": {
|
| 26 |
+
"_target_": "torch.nn.CrossEntropyLoss"
|
| 27 |
+
},
|
| 28 |
+
"optimizer": {
|
| 29 |
+
"_target_": "torch.optim.Adam",
|
| 30 |
+
"params": "[email protected]()",
|
| 31 |
+
"lr": 0.0001,
|
| 32 |
+
"weight_decay": 1e-05
|
| 33 |
+
},
|
| 34 |
+
"lr_scheduler": {
|
| 35 |
+
"_target_": "torch.optim.lr_scheduler.StepLR",
|
| 36 |
+
"optimizer": "@optimizer",
|
| 37 |
+
"step_size": 50
|
| 38 |
+
},
|
| 39 |
+
"max_epochs": 100,
|
| 40 |
+
"train": {
|
| 41 |
+
"preprocessing": {
|
| 42 |
+
"_target_": "Compose",
|
| 43 |
+
"transforms": [
|
| 44 |
+
{
|
| 45 |
+
"_target_": "LoadImaged",
|
| 46 |
+
"keys": [
|
| 47 |
+
"image",
|
| 48 |
+
"label"
|
| 49 |
+
],
|
| 50 |
+
"dtype": "uint8",
|
| 51 |
+
"image_only": false
|
| 52 |
+
},
|
| 53 |
+
{
|
| 54 |
+
"_target_": "EnsureChannelFirstd",
|
| 55 |
+
"keys": [
|
| 56 |
+
"image",
|
| 57 |
+
"label"
|
| 58 |
+
]
|
| 59 |
+
},
|
| 60 |
+
{
|
| 61 |
+
"_target_": "SplitLabeld",
|
| 62 |
+
"keys": "label",
|
| 63 |
+
"mask_value": "",
|
| 64 |
+
"others_value": 255,
|
| 65 |
+
"to_binary_mask": false
|
| 66 |
+
},
|
| 67 |
+
{
|
| 68 |
+
"_target_": "RandTorchVisiond",
|
| 69 |
+
"keys": "image",
|
| 70 |
+
"name": "ColorJitter",
|
| 71 |
+
"brightness": 0.25,
|
| 72 |
+
"contrast": 0.75,
|
| 73 |
+
"saturation": 0.25,
|
| 74 |
+
"hue": 0.04
|
| 75 |
+
},
|
| 76 |
+
{
|
| 77 |
+
"_target_": "RandFlipd",
|
| 78 |
+
"keys": [
|
| 79 |
+
"image",
|
| 80 |
+
"label",
|
| 81 |
+
"others"
|
| 82 |
+
],
|
| 83 |
+
"prob": 0.5
|
| 84 |
+
},
|
| 85 |
+
{
|
| 86 |
+
"_target_": "RandRotate90d",
|
| 87 |
+
"keys": [
|
| 88 |
+
"image",
|
| 89 |
+
"label",
|
| 90 |
+
"others"
|
| 91 |
+
],
|
| 92 |
+
"prob": 0.5
|
| 93 |
+
},
|
| 94 |
+
{
|
| 95 |
+
"_target_": "ScaleIntensityRanged",
|
| 96 |
+
"keys": "image",
|
| 97 |
+
"a_min": 0.0,
|
| 98 |
+
"a_max": 255.0,
|
| 99 |
+
"b_min": -1.0,
|
| 100 |
+
"b_max": 1.0
|
| 101 |
+
},
|
| 102 |
+
{
|
| 103 |
+
"_target_": "AddLabelAsGuidanced",
|
| 104 |
+
"keys": "image",
|
| 105 |
+
"source": "label"
|
| 106 |
+
},
|
| 107 |
+
{
|
| 108 |
+
"_target_": "SetLabelClassd",
|
| 109 |
+
"keys": "label",
|
| 110 |
+
"offset": -1
|
| 111 |
+
},
|
| 112 |
+
{
|
| 113 |
+
"_target_": "SelectItemsd",
|
| 114 |
+
"keys": [
|
| 115 |
+
"image",
|
| 116 |
+
"label"
|
| 117 |
+
]
|
| 118 |
+
}
|
| 119 |
+
]
|
| 120 |
+
},
|
| 121 |
+
"dataset": {
|
| 122 |
+
"_target_": "CacheDataset",
|
| 123 |
+
"data": "@train_datalist",
|
| 124 |
+
"transform": "@train#preprocessing",
|
| 125 |
+
"cache_rate": 1.0,
|
| 126 |
+
"num_workers": 4
|
| 127 |
+
},
|
| 128 |
+
"dataloader": {
|
| 129 |
+
"_target_": "DataLoader",
|
| 130 |
+
"dataset": "@train#dataset",
|
| 131 |
+
"batch_size": 64,
|
| 132 |
+
"shuffle": true,
|
| 133 |
+
"num_workers": 4
|
| 134 |
+
},
|
| 135 |
+
"inferer": {
|
| 136 |
+
"_target_": "SimpleInferer"
|
| 137 |
+
},
|
| 138 |
+
"postprocessing": {
|
| 139 |
+
"_target_": "Compose",
|
| 140 |
+
"transforms": [
|
| 141 |
+
{
|
| 142 |
+
"_target_": "Activationsd",
|
| 143 |
+
"keys": "pred",
|
| 144 |
+
"softmax": true
|
| 145 |
+
},
|
| 146 |
+
{
|
| 147 |
+
"_target_": "AsDiscreted",
|
| 148 |
+
"keys": [
|
| 149 |
+
"pred",
|
| 150 |
+
"label"
|
| 151 |
+
],
|
| 152 |
+
"argmax": [
|
| 153 |
+
true,
|
| 154 |
+
false
|
| 155 |
+
],
|
| 156 |
+
"to_onehot": 4
|
| 157 |
+
},
|
| 158 |
+
{
|
| 159 |
+
"_target_": "ToTensord",
|
| 160 |
+
"keys": [
|
| 161 |
+
"pred",
|
| 162 |
+
"label"
|
| 163 |
+
],
|
| 164 |
+
"device": "@device"
|
| 165 |
+
}
|
| 166 |
+
]
|
| 167 |
+
},
|
| 168 |
+
"handlers": [
|
| 169 |
+
{
|
| 170 |
+
"_target_": "LrScheduleHandler",
|
| 171 |
+
"lr_scheduler": "@lr_scheduler",
|
| 172 |
+
"print_lr": true
|
| 173 |
+
},
|
| 174 |
+
{
|
| 175 |
+
"_target_": "ValidationHandler",
|
| 176 |
+
"validator": "@validate#evaluator",
|
| 177 |
+
"epoch_level": true,
|
| 178 |
+
"interval": "@val_interval"
|
| 179 |
+
},
|
| 180 |
+
{
|
| 181 |
+
"_target_": "StatsHandler",
|
| 182 |
+
"tag_name": "train_loss",
|
| 183 |
+
"output_transform": "$monai.handlers.from_engine(['loss'], first=True)"
|
| 184 |
+
},
|
| 185 |
+
{
|
| 186 |
+
"_target_": "TensorBoardStatsHandler",
|
| 187 |
+
"log_dir": "@output_dir",
|
| 188 |
+
"tag_name": "train_loss",
|
| 189 |
+
"output_transform": "$monai.handlers.from_engine(['loss'], first=True)"
|
| 190 |
+
},
|
| 191 |
+
{
|
| 192 |
+
"_target_": "scripts.TensorBoardImageHandler",
|
| 193 |
+
"class_names": {
|
| 194 |
+
"0": "Other",
|
| 195 |
+
"1": "Inflammatory",
|
| 196 |
+
"2": "Epithelial",
|
| 197 |
+
"3": "Spindle-Shaped"
|
| 198 |
+
},
|
| 199 |
+
"log_dir": "@output_dir",
|
| 200 |
+
"batch_limit": 4,
|
| 201 |
+
"tag_name": "train"
|
| 202 |
+
}
|
| 203 |
+
],
|
| 204 |
+
"key_metric": {
|
| 205 |
+
"train_f1": {
|
| 206 |
+
"_target_": "ConfusionMatrix",
|
| 207 |
+
"metric_name": "f1 score",
|
| 208 |
+
"output_transform": "$monai.handlers.from_engine(['pred', 'label'])"
|
| 209 |
+
}
|
| 210 |
+
},
|
| 211 |
+
"trainer": {
|
| 212 |
+
"_target_": "SupervisedTrainer",
|
| 213 |
+
"max_epochs": "@max_epochs",
|
| 214 |
+
"device": "@device",
|
| 215 |
+
"train_data_loader": "@train#dataloader",
|
| 216 |
+
"network": "@network",
|
| 217 |
+
"loss_function": "@loss",
|
| 218 |
+
"optimizer": "@optimizer",
|
| 219 |
+
"inferer": "@train#inferer",
|
| 220 |
+
"postprocessing": "@train#postprocessing",
|
| 221 |
+
"key_train_metric": "@train#key_metric",
|
| 222 |
+
"train_handlers": "@train#handlers",
|
| 223 |
+
"amp": true
|
| 224 |
+
}
|
| 225 |
+
},
|
| 226 |
+
"validate": {
|
| 227 |
+
"preprocessing": {
|
| 228 |
+
"_target_": "Compose",
|
| 229 |
+
"transforms": [
|
| 230 |
+
{
|
| 231 |
+
"_target_": "LoadImaged",
|
| 232 |
+
"keys": [
|
| 233 |
+
"image",
|
| 234 |
+
"label"
|
| 235 |
+
],
|
| 236 |
+
"dtype": "uint8",
|
| 237 |
+
"image_only": false
|
| 238 |
+
},
|
| 239 |
+
{
|
| 240 |
+
"_target_": "EnsureChannelFirstd",
|
| 241 |
+
"keys": [
|
| 242 |
+
"image",
|
| 243 |
+
"label"
|
| 244 |
+
]
|
| 245 |
+
},
|
| 246 |
+
{
|
| 247 |
+
"_target_": "SplitLabeld",
|
| 248 |
+
"keys": "label",
|
| 249 |
+
"mask_value": "",
|
| 250 |
+
"others_value": 255,
|
| 251 |
+
"to_binary_mask": false
|
| 252 |
+
},
|
| 253 |
+
{
|
| 254 |
+
"_target_": "ScaleIntensityRanged",
|
| 255 |
+
"keys": "image",
|
| 256 |
+
"a_min": 0.0,
|
| 257 |
+
"a_max": 255.0,
|
| 258 |
+
"b_min": -1.0,
|
| 259 |
+
"b_max": 1.0
|
| 260 |
+
},
|
| 261 |
+
{
|
| 262 |
+
"_target_": "AddLabelAsGuidanced",
|
| 263 |
+
"keys": "image",
|
| 264 |
+
"source": "label"
|
| 265 |
+
},
|
| 266 |
+
{
|
| 267 |
+
"_target_": "SetLabelClassd",
|
| 268 |
+
"keys": "label",
|
| 269 |
+
"offset": -1
|
| 270 |
+
},
|
| 271 |
+
{
|
| 272 |
+
"_target_": "SelectItemsd",
|
| 273 |
+
"keys": [
|
| 274 |
+
"image",
|
| 275 |
+
"label",
|
| 276 |
+
"image_meta_dict"
|
| 277 |
+
]
|
| 278 |
+
}
|
| 279 |
+
]
|
| 280 |
+
},
|
| 281 |
+
"dataset": {
|
| 282 |
+
"_target_": "CacheDataset",
|
| 283 |
+
"data": "@val_datalist",
|
| 284 |
+
"transform": "@validate#preprocessing",
|
| 285 |
+
"cache_rate": 1.0
|
| 286 |
+
},
|
| 287 |
+
"dataloader": {
|
| 288 |
+
"_target_": "DataLoader",
|
| 289 |
+
"dataset": "@validate#dataset",
|
| 290 |
+
"batch_size": 64,
|
| 291 |
+
"shuffle": false,
|
| 292 |
+
"num_workers": 4
|
| 293 |
+
},
|
| 294 |
+
"inferer": {
|
| 295 |
+
"_target_": "SimpleInferer"
|
| 296 |
+
},
|
| 297 |
+
"postprocessing": "%train#postprocessing",
|
| 298 |
+
"handlers": [
|
| 299 |
+
{
|
| 300 |
+
"_target_": "StatsHandler",
|
| 301 |
+
"iteration_log": false
|
| 302 |
+
},
|
| 303 |
+
{
|
| 304 |
+
"_target_": "TensorBoardStatsHandler",
|
| 305 |
+
"log_dir": "@output_dir",
|
| 306 |
+
"iteration_log": false
|
| 307 |
+
},
|
| 308 |
+
{
|
| 309 |
+
"_target_": "CheckpointSaver",
|
| 310 |
+
"save_dir": "@ckpt_dir",
|
| 311 |
+
"save_dict": {
|
| 312 |
+
"model": "@network"
|
| 313 |
+
},
|
| 314 |
+
"save_key_metric": true,
|
| 315 |
+
"key_metric_filename": "model.pt"
|
| 316 |
+
},
|
| 317 |
+
{
|
| 318 |
+
"_target_": "scripts.TensorBoardImageHandler",
|
| 319 |
+
"class_names": {
|
| 320 |
+
"0": "Other",
|
| 321 |
+
"1": "Inflammatory",
|
| 322 |
+
"2": "Epithelial",
|
| 323 |
+
"3": "Spindle-Shaped"
|
| 324 |
+
},
|
| 325 |
+
"log_dir": "@output_dir",
|
| 326 |
+
"batch_limit": 8,
|
| 327 |
+
"tag_name": "val"
|
| 328 |
+
}
|
| 329 |
+
],
|
| 330 |
+
"key_metric": {
|
| 331 |
+
"val_f1": {
|
| 332 |
+
"_target_": "ConfusionMatrix",
|
| 333 |
+
"metric_name": "f1 score",
|
| 334 |
+
"output_transform": "$monai.handlers.from_engine(['pred', 'label'])"
|
| 335 |
+
}
|
| 336 |
+
},
|
| 337 |
+
"additional_metrics": {
|
| 338 |
+
"val_accuracy": {
|
| 339 |
+
"_target_": "ignite.metrics.Accuracy",
|
| 340 |
+
"output_transform": "$monai.handlers.from_engine(['pred', 'label'])"
|
| 341 |
+
}
|
| 342 |
+
},
|
| 343 |
+
"evaluator": {
|
| 344 |
+
"_target_": "SupervisedEvaluator",
|
| 345 |
+
"device": "@device",
|
| 346 |
+
"val_data_loader": "@validate#dataloader",
|
| 347 |
+
"network": "@network",
|
| 348 |
+
"inferer": "@validate#inferer",
|
| 349 |
+
"postprocessing": "@validate#postprocessing",
|
| 350 |
+
"key_val_metric": "@validate#key_metric",
|
| 351 |
+
"additional_metrics": "@validate#additional_metrics",
|
| 352 |
+
"val_handlers": "@validate#handlers",
|
| 353 |
+
"amp": true
|
| 354 |
+
}
|
| 355 |
+
},
|
| 356 |
+
"initialize": [
|
| 357 |
+
"$import sys",
|
| 358 |
+
"$sys.path.append(@bundle_root)",
|
| 359 |
+
"$monai.utils.set_determinism(seed=123)"
|
| 360 |
+
],
|
| 361 |
+
"run": [
|
| 362 |
+
"$@train#trainer.run()"
|
| 363 |
+
]
|
| 364 |
+
}
|
docs/README.md
ADDED
|
@@ -0,0 +1,232 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
# Model Overview
|
| 2 |
+
A pre-trained model for classifying nuclei cells as the following types
|
| 3 |
+
- Other
|
| 4 |
+
- Inflammatory
|
| 5 |
+
- Epithelial
|
| 6 |
+
- Spindle-Shaped
|
| 7 |
+
|
| 8 |
+
This model is trained using [DenseNet121](https://docs.monai.io/en/latest/networks.html#densenet121) over [ConSeP](https://warwick.ac.uk/fac/cross_fac/tia/data/hovernet) dataset.
|
| 9 |
+
|
| 10 |
+
## Data
|
| 11 |
+
The training dataset is from https://warwick.ac.uk/fac/cross_fac/tia/data/hovernet
|
| 12 |
+
```commandline
|
| 13 |
+
wget https://warwick.ac.uk/fac/cross_fac/tia/data/hovernet/consep_dataset.zip
|
| 14 |
+
unzip -q consep_dataset.zip
|
| 15 |
+
```
|
| 16 |
+
<br/>
|
| 17 |
+
|
| 18 |
+
### Preprocessing
|
| 19 |
+
After [downloading this dataset](https://warwick.ac.uk/fac/cross_fac/tia/data/hovernet/consep_dataset.zip),
|
| 20 |
+
python script `data_process.py` from `scripts` folder can be used to preprocess and generate the final dataset for training.
|
| 21 |
+
|
| 22 |
+
```commandline
|
| 23 |
+
python scripts/data_process.py --input /path/to/data/CoNSeP --output /path/to/data/CoNSePNuclei
|
| 24 |
+
```
|
| 25 |
+
|
| 26 |
+
After generating the output files, please modify the `dataset_dir` parameter specified in `configs/train.json` and `configs/inference.json` to reflect the output folder which contains new dataset.json.
|
| 27 |
+
|
| 28 |
+
Class values in dataset are
|
| 29 |
+
|
| 30 |
+
- 1 = other
|
| 31 |
+
- 2 = inflammatory
|
| 32 |
+
- 3 = healthy epithelial
|
| 33 |
+
- 4 = dysplastic/malignant epithelial
|
| 34 |
+
- 5 = fibroblast
|
| 35 |
+
- 6 = muscle
|
| 36 |
+
- 7 = endothelial
|
| 37 |
+
|
| 38 |
+
As part of pre-processing, the following steps are executed.
|
| 39 |
+
|
| 40 |
+
- Crop and Extract each nuclei Image + Label (128x128) based on the centroid given in the dataset.
|
| 41 |
+
- Combine classes 3 & 4 into the epithelial class and 5,6 & 7 into the spindle-shaped class.
|
| 42 |
+
- Update the label index for the target nuclie based on the class value
|
| 43 |
+
- Other cells which are part of the patch are modified to have label idex = 255
|
| 44 |
+
|
| 45 |
+
Example `dataset.json` in output folder:
|
| 46 |
+
```json
|
| 47 |
+
{
|
| 48 |
+
"training": [
|
| 49 |
+
{
|
| 50 |
+
"image": "/workspace/data/CoNSePNuclei/Train/Images/train_1_3_0001.png",
|
| 51 |
+
"label": "/workspace/data/CoNSePNuclei/Train/Labels/train_1_3_0001.png",
|
| 52 |
+
"nuclei_id": 1,
|
| 53 |
+
"mask_value": 3,
|
| 54 |
+
"centroid": [
|
| 55 |
+
64,
|
| 56 |
+
64
|
| 57 |
+
]
|
| 58 |
+
}
|
| 59 |
+
],
|
| 60 |
+
"validation": [
|
| 61 |
+
{
|
| 62 |
+
"image": "/workspace/data/CoNSePNuclei/Test/Images/test_1_3_0001.png",
|
| 63 |
+
"label": "/workspace/data/CoNSePNuclei/Test/Labels/test_1_3_0001.png",
|
| 64 |
+
"nuclei_id": 1,
|
| 65 |
+
"mask_value": 3,
|
| 66 |
+
"centroid": [
|
| 67 |
+
64,
|
| 68 |
+
64
|
| 69 |
+
]
|
| 70 |
+
}
|
| 71 |
+
]
|
| 72 |
+
}
|
| 73 |
+
```
|
| 74 |
+
|
| 75 |
+
## Training configuration
|
| 76 |
+
The training was performed with the following:
|
| 77 |
+
|
| 78 |
+
- GPU: at least 12GB of GPU memory
|
| 79 |
+
- Actual Model Input: 4 x 128 x 128
|
| 80 |
+
- AMP: True
|
| 81 |
+
- Optimizer: Adam
|
| 82 |
+
- Learning Rate: 1e-4
|
| 83 |
+
- Loss: torch.nn.CrossEntropyLoss
|
| 84 |
+
- Dataset Manager: CacheDataset
|
| 85 |
+
|
| 86 |
+
### Memory Consumption Warning
|
| 87 |
+
|
| 88 |
+
If you face memory issues with CacheDataset, you can either switch to a regular Dataset class or lower the caching rate `cache_rate` in the configurations within range [0, 1] to minimize the System RAM requirements.
|
| 89 |
+
|
| 90 |
+
## Input
|
| 91 |
+
4 channels
|
| 92 |
+
- 3 RGB channels
|
| 93 |
+
- 1 signal channel (label mask)
|
| 94 |
+
|
| 95 |
+
## Output
|
| 96 |
+
4 channels
|
| 97 |
+
- 0 = Other
|
| 98 |
+
- 1 = Inflammatory
|
| 99 |
+
- 2 = Epithelial
|
| 100 |
+
- 3 = Spindle-Shaped
|
| 101 |
+
|
| 102 |
+
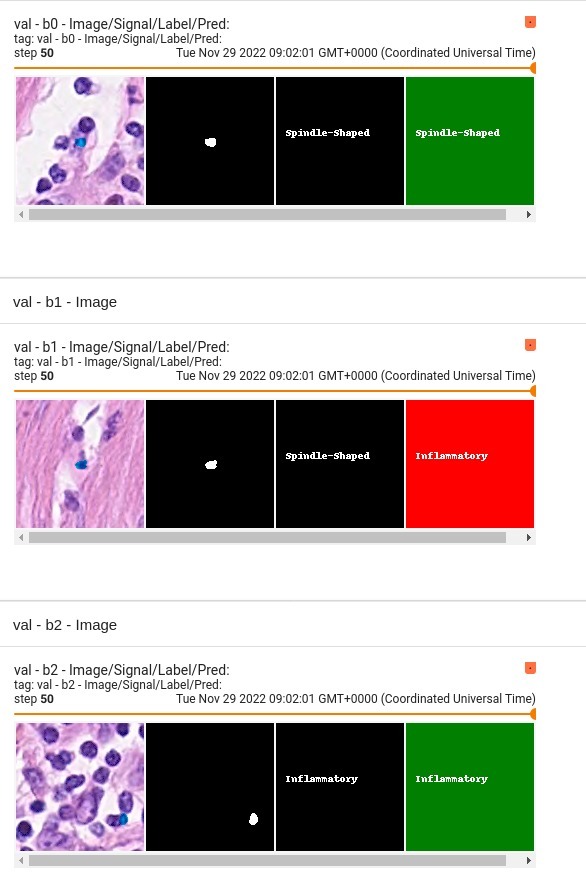
|
| 103 |
+
|
| 104 |
+
## Performance
|
| 105 |
+
This model achieves the following F1 score on the validation data provided as part of the dataset:
|
| 106 |
+
|
| 107 |
+
- Train F1 score = 0.926
|
| 108 |
+
- Validation F1 score = 0.852
|
| 109 |
+
|
| 110 |
+
<hr/>
|
| 111 |
+
Confusion Metrics for <b>Validation</b> for individual classes are:
|
| 112 |
+
|
| 113 |
+
| Metric | Other | Inflammatory | Epithelial | Spindle-Shaped |
|
| 114 |
+
|-----------|--------|--------------|------------|----------------|
|
| 115 |
+
| Precision | 0.6909 | 0.7773 | 0.9078 | 0.8478 |
|
| 116 |
+
| Recall | 0.2754 | 0.7831 | 0.9533 | 0.8514 |
|
| 117 |
+
| F1-score | 0.3938 | 0.7802 | 0.9300 | 0.8496 |
|
| 118 |
+
|
| 119 |
+
|
| 120 |
+
<hr/>
|
| 121 |
+
Confusion Metrics for <b>Training</b> for individual classes are:
|
| 122 |
+
|
| 123 |
+
| Metric | Other | Inflammatory | Epithelial | Spindle-Shaped |
|
| 124 |
+
|-----------|--------|--------------|------------|----------------|
|
| 125 |
+
| Precision | 0.8000 | 0.9076 | 0.9560 | 0.9019 |
|
| 126 |
+
| Recall | 0.6512 | 0.9028 | 0.9690 | 0.8989 |
|
| 127 |
+
| F1-score | 0.7179 | 0.9052 | 0.9625 | 0.9004 |
|
| 128 |
+
|
| 129 |
+
|
| 130 |
+
|
| 131 |
+
#### Training Loss and F1
|
| 132 |
+
A graph showing the training Loss and F1-score over 100 epochs.
|
| 133 |
+
|
| 134 |
+
 <br>
|
| 135 |
+
 <br>
|
| 136 |
+
|
| 137 |
+
#### Validation F1
|
| 138 |
+
A graph showing the validation F1-score over 100 epochs.
|
| 139 |
+
|
| 140 |
+
 <br>
|
| 141 |
+
|
| 142 |
+
#### TensorRT speedup
|
| 143 |
+
This bundle supports acceleration with TensorRT. The table below displays the speedup ratios observed on an A100 80G GPU. Please note that 32-bit precision models are benchmarked with tf32 weight format.
|
| 144 |
+
|
| 145 |
+
| method | torch_tf32(ms) | torch_amp(ms) | trt_tf32(ms) | trt_fp16(ms) | speedup amp | speedup tf32 | speedup fp16 | amp vs fp16|
|
| 146 |
+
| :---: | :---: | :---: | :---: | :---: | :---: | :---: | :---: | :---: |
|
| 147 |
+
| model computation | 20.47 | 20.57 | 2.49 | 1.48 | 1.00 | 8.22 | 13.83 | 13.90 |
|
| 148 |
+
| end2end | 45 | 49 | 18 | 18 | 0.92 | 2.50 | 2.50 | 2.72 |
|
| 149 |
+
|
| 150 |
+
Where:
|
| 151 |
+
- `model computation` means the speedup ratio of model's inference with a random input without preprocessing and postprocessing
|
| 152 |
+
- `end2end` means run the bundle end-to-end with the TensorRT based model.
|
| 153 |
+
- `torch_tf32` and `torch_amp` are for the PyTorch models with or without `amp` mode.
|
| 154 |
+
- `trt_tf32` and `trt_fp16` are for the TensorRT based models converted in corresponding precision.
|
| 155 |
+
- `speedup amp`, `speedup tf32` and `speedup fp16` are the speedup ratios of corresponding models versus the PyTorch float32 model
|
| 156 |
+
- `amp vs fp16` is the speedup ratio between the PyTorch amp model and the TensorRT float16 based model.
|
| 157 |
+
|
| 158 |
+
This result is benchmarked under:
|
| 159 |
+
- TensorRT: 10.3.0+cuda12.6
|
| 160 |
+
- Torch-TensorRT Version: 2.4.0
|
| 161 |
+
- CPU Architecture: x86-64
|
| 162 |
+
- OS: ubuntu 20.04
|
| 163 |
+
- Python version:3.10.12
|
| 164 |
+
- CUDA version: 12.6
|
| 165 |
+
- GPU models and configuration: A100 80G
|
| 166 |
+
|
| 167 |
+
## MONAI Bundle Commands
|
| 168 |
+
In addition to the Pythonic APIs, a few command line interfaces (CLI) are provided to interact with the bundle. The CLI supports flexible use cases, such as overriding configs at runtime and predefining arguments in a file.
|
| 169 |
+
|
| 170 |
+
For more details usage instructions, visit the [MONAI Bundle Configuration Page](https://docs.monai.io/en/latest/config_syntax.html).
|
| 171 |
+
|
| 172 |
+
#### Execute training:
|
| 173 |
+
|
| 174 |
+
```
|
| 175 |
+
python -m monai.bundle run --config_file configs/train.json
|
| 176 |
+
```
|
| 177 |
+
|
| 178 |
+
Please note that if the default dataset path is not modified with the actual path in the bundle config files, you can also override it by using `--dataset_dir`:
|
| 179 |
+
|
| 180 |
+
```
|
| 181 |
+
python -m monai.bundle run --config_file configs/train.json --dataset_dir <actual dataset path>
|
| 182 |
+
```
|
| 183 |
+
|
| 184 |
+
#### Override the `train` config to execute multi-GPU training:
|
| 185 |
+
|
| 186 |
+
```
|
| 187 |
+
torchrun --standalone --nnodes=1 --nproc_per_node=2 -m monai.bundle run --config_file "['configs/train.json','configs/multi_gpu_train.json']"
|
| 188 |
+
```
|
| 189 |
+
|
| 190 |
+
Please note that the distributed training-related options depend on the actual running environment; thus, users may need to remove `--standalone`, modify `--nnodes`, or do some other necessary changes according to the machine used. For more details, please refer to [pytorch's official tutorial](https://pytorch.org/tutorials/intermediate/ddp_tutorial.html).
|
| 191 |
+
|
| 192 |
+
#### Override the `train` config to execute evaluation with the trained model:
|
| 193 |
+
|
| 194 |
+
```
|
| 195 |
+
python -m monai.bundle run --config_file "['configs/train.json','configs/evaluate.json']"
|
| 196 |
+
```
|
| 197 |
+
|
| 198 |
+
#### Override the `train` config and `evaluate` config to execute multi-GPU evaluation:
|
| 199 |
+
|
| 200 |
+
```
|
| 201 |
+
torchrun --standalone --nnodes=1 --nproc_per_node=2 -m monai.bundle run --config_file "['configs/train.json','configs/evaluate.json','configs/multi_gpu_evaluate.json']"
|
| 202 |
+
```
|
| 203 |
+
|
| 204 |
+
#### Execute inference:
|
| 205 |
+
|
| 206 |
+
```
|
| 207 |
+
python -m monai.bundle run --config_file configs/inference.json
|
| 208 |
+
```
|
| 209 |
+
|
| 210 |
+
#### Execute inference with the TensorRT model:
|
| 211 |
+
|
| 212 |
+
```
|
| 213 |
+
python -m monai.bundle run --config_file "['configs/inference.json', 'configs/inference_trt.json']"
|
| 214 |
+
```
|
| 215 |
+
|
| 216 |
+
# References
|
| 217 |
+
[1] S. Graham, Q. D. Vu, S. E. A. Raza, A. Azam, Y-W. Tsang, J. T. Kwak and N. Rajpoot. "HoVer-Net: Simultaneous Segmentation and Classification of Nuclei in Multi-Tissue Histology Images." Medical Image Analysis, Sept. 2019. [[doi](https://doi.org/10.1016/j.media.2019.101563)]
|
| 218 |
+
|
| 219 |
+
# License
|
| 220 |
+
Copyright (c) MONAI Consortium
|
| 221 |
+
|
| 222 |
+
Licensed under the Apache License, Version 2.0 (the "License");
|
| 223 |
+
you may not use this file except in compliance with the License.
|
| 224 |
+
You may obtain a copy of the License at
|
| 225 |
+
|
| 226 |
+
http://www.apache.org/licenses/LICENSE-2.0
|
| 227 |
+
|
| 228 |
+
Unless required by applicable law or agreed to in writing, software
|
| 229 |
+
distributed under the License is distributed on an "AS IS" BASIS,
|
| 230 |
+
WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.
|
| 231 |
+
See the License for the specific language governing permissions and
|
| 232 |
+
limitations under the License.
|
docs/data_license.txt
ADDED
|
@@ -0,0 +1,6 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
Third Party Licenses
|
| 2 |
+
-----------------------------------------------------------------------
|
| 3 |
+
|
| 4 |
+
/*********************************************************************/
|
| 5 |
+
i. HoVer-Net: Simultaneous Segmentation and Classification of Nuclei in Multi-Tissue Histology Images
|
| 6 |
+
https://warwick.ac.uk/fac/cross_fac/tia/data/hovernet/
|
models/model.pt
ADDED
|
@@ -0,0 +1,3 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
version https://git-lfs.github.com/spec/v1
|
| 2 |
+
oid sha256:54066a13f090b94eb43c589c9090f9791810f74903998418c20e8502839d2c83
|
| 3 |
+
size 28419489
|
models/model.ts
ADDED
|
@@ -0,0 +1,3 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
version https://git-lfs.github.com/spec/v1
|
| 2 |
+
oid sha256:10745ecb9c680c3bc2756da88326403153eee0964551a62da884d5fb21116f5e
|
| 3 |
+
size 28573133
|
scripts/__init__.py
ADDED
|
@@ -0,0 +1,13 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
# Copyright (c) MONAI Consortium
|
| 2 |
+
# Licensed under the Apache License, Version 2.0 (the "License");
|
| 3 |
+
# you may not use this file except in compliance with the License.
|
| 4 |
+
# You may obtain a copy of the License at
|
| 5 |
+
# http://www.apache.org/licenses/LICENSE-2.0
|
| 6 |
+
# Unless required by applicable law or agreed to in writing, software
|
| 7 |
+
# distributed under the License is distributed on an "AS IS" BASIS,
|
| 8 |
+
# WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.
|
| 9 |
+
# See the License for the specific language governing permissions and
|
| 10 |
+
# limitations under the License.
|
| 11 |
+
|
| 12 |
+
from .handlers import TensorBoardImageHandler
|
| 13 |
+
from .writer import ClassificationWriter
|
scripts/data_process.py
ADDED
|
@@ -0,0 +1,79 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
# Copyright (c) MONAI Consortium
|
| 2 |
+
# Licensed under the Apache License, Version 2.0 (the "License");
|
| 3 |
+
# you may not use this file except in compliance with the License.
|
| 4 |
+
# You may obtain a copy of the License at
|
| 5 |
+
# http://www.apache.org/licenses/LICENSE-2.0
|
| 6 |
+
# Unless required by applicable law or agreed to in writing, software
|
| 7 |
+
# distributed under the License is distributed on an "AS IS" BASIS,
|
| 8 |
+
# WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.
|
| 9 |
+
# See the License for the specific language governing permissions and
|
| 10 |
+
# limitations under the License.
|
| 11 |
+
import argparse
|
| 12 |
+
import glob
|
| 13 |
+
import json
|
| 14 |
+
import logging
|
| 15 |
+
import os
|
| 16 |
+
|
| 17 |
+
from dataset import consep_nuclei_dataset
|
| 18 |
+
|
| 19 |
+
logger = logging.getLogger(__name__)
|
| 20 |
+
|
| 21 |
+
|
| 22 |
+
def main():
|
| 23 |
+
logging.basicConfig(
|
| 24 |
+
level=logging.INFO,
|
| 25 |
+
format="[%(asctime)s] [%(process)s] [%(threadName)s] [%(levelname)s] (%(name)s:%(lineno)d) - %(message)s",
|
| 26 |
+
datefmt="%Y-%m-%d %H:%M:%S",
|
| 27 |
+
force=True,
|
| 28 |
+
)
|
| 29 |
+
|
| 30 |
+
parser = argparse.ArgumentParser()
|
| 31 |
+
parser.add_argument(
|
| 32 |
+
"--input",
|
| 33 |
+
"-i",
|
| 34 |
+
type=str,
|
| 35 |
+
default=r"/workspace/data/CoNSeP",
|
| 36 |
+
help="Input/Downloaded/Extracted dir for CoNSeP Dataset",
|
| 37 |
+
)
|
| 38 |
+
parser.add_argument(
|
| 39 |
+
"--output",
|
| 40 |
+
"-o",
|
| 41 |
+
type=str,
|
| 42 |
+
default=r"/workspace/data/CoNSePNuclei",
|
| 43 |
+
help="Output dir to store pre-processed data",
|
| 44 |
+
)
|
| 45 |
+
parser.add_argument("--crop_size", "-s", type=int, default=128, help="Crop size for each Nuclei")
|
| 46 |
+
parser.add_argument("--limit", "-n", type=int, default=0, help="Non-zero value to limit processing max records")
|
| 47 |
+
|
| 48 |
+
args = parser.parse_args()
|
| 49 |
+
dataset_json = {}
|
| 50 |
+
for f, v in {"Train": "training", "Test": "validation"}.items():
|
| 51 |
+
logger.info("---------------------------------------------------------------------------------")
|
| 52 |
+
if not os.path.exists(os.path.join(args.input, f)):
|
| 53 |
+
logger.warning(f"Ignore {f} (NOT Exists in Input Folder)")
|
| 54 |
+
continue
|
| 55 |
+
|
| 56 |
+
logger.info(f"Processing Images/labels for: {f}")
|
| 57 |
+
images_path = os.path.join(args.input, f, "Images", "*.png")
|
| 58 |
+
labels_path = os.path.join(args.input, f, "Labels", "*.mat")
|
| 59 |
+
images = sorted(glob.glob(images_path))
|
| 60 |
+
labels = sorted(glob.glob(labels_path))
|
| 61 |
+
ds = [{"image": i, "label": l} for i, l in zip(images, labels)]
|
| 62 |
+
|
| 63 |
+
output_dir = os.path.join(args.output, f) if args.output else f
|
| 64 |
+
crop_size = args.crop_size
|
| 65 |
+
limit = args.limit
|
| 66 |
+
|
| 67 |
+
ds_new = consep_nuclei_dataset(ds, output_dir, crop_size, limit=limit)
|
| 68 |
+
logger.info(f"Total Generated/Extended Records: {len(ds)} => {len(ds_new)}")
|
| 69 |
+
|
| 70 |
+
dataset_json[v] = ds_new
|
| 71 |
+
|
| 72 |
+
ds_file = os.path.join(args.output, "dataset.json")
|
| 73 |
+
with open(ds_file, "w") as fp:
|
| 74 |
+
json.dump(dataset_json, fp, indent=2)
|
| 75 |
+
logger.info(f"Dataset JSON Generated at: {ds_file}")
|
| 76 |
+
|
| 77 |
+
|
| 78 |
+
if __name__ == "__main__":
|
| 79 |
+
main()
|
scripts/dataset.py
ADDED
|
@@ -0,0 +1,189 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
# Copyright (c) MONAI Consortium
|
| 2 |
+
# Licensed under the Apache License, Version 2.0 (the "License");
|
| 3 |
+
# you may not use this file except in compliance with the License.
|
| 4 |
+
# You may obtain a copy of the License at
|
| 5 |
+
# http://www.apache.org/licenses/LICENSE-2.0
|
| 6 |
+
# Unless required by applicable law or agreed to in writing, software
|
| 7 |
+
# distributed under the License is distributed on an "AS IS" BASIS,
|
| 8 |
+
# WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.
|
| 9 |
+
# See the License for the specific language governing permissions and
|
| 10 |
+
# limitations under the License.
|
| 11 |
+
|
| 12 |
+
import copy
|
| 13 |
+
import json
|
| 14 |
+
import logging
|
| 15 |
+
import os
|
| 16 |
+
import pathlib
|
| 17 |
+
from typing import Dict, List
|
| 18 |
+
|
| 19 |
+
import numpy as np
|
| 20 |
+
from monai.apps.utils import tqdm
|
| 21 |
+
from monai.utils import optional_import
|
| 22 |
+
|
| 23 |
+
loadmat, _ = optional_import("scipy.io", name="loadmat")
|
| 24 |
+
PILImage, _ = optional_import("PIL.Image")
|
| 25 |
+
|
| 26 |
+
|
| 27 |
+
def consep_nuclei_dataset(datalist, output_dir, crop_size, min_area=80, min_distance=20, limit=0) -> List[Dict]:
|
| 28 |
+
"""
|
| 29 |
+
Utility to pre-process and create dataset list for Patches per Nuclei for training over ConSeP dataset.
|
| 30 |
+
|
| 31 |
+
Args:
|
| 32 |
+
datalist: A list of data dictionary. Each entry should at least contain 'image_key': <image filename>.
|
| 33 |
+
For example, typical input data can be a list of dictionaries::
|
| 34 |
+
|
| 35 |
+
[{'image': <image filename>, 'label': <label filename>}]
|
| 36 |
+
|
| 37 |
+
output_dir: target directory to store the training data after flattening
|
| 38 |
+
crop_size: Crop Size for each patch
|
| 39 |
+
min_area: Min Area for each nuclei to be included in dataset
|
| 40 |
+
min_distance: Min Distance from boundary for each nuclei to be included in dataset
|
| 41 |
+
limit: limit number of inputs for pre-processing. Defaults to 0 (no limit).
|
| 42 |
+
|
| 43 |
+
Raises:
|
| 44 |
+
ValueError: When ``datalist`` is Empty
|
| 45 |
+
ValueError: When ``scipy.io.loadmat`` is Not available
|
| 46 |
+
|
| 47 |
+
Returns:
|
| 48 |
+
A new datalist that contains path to the images/labels after pre-processing.
|
| 49 |
+
|
| 50 |
+
Example::
|
| 51 |
+
|
| 52 |
+
datalist = consep_nuclei_dataset(
|
| 53 |
+
datalist=[{'image': 'img1.png', 'label': 'label1.mat'}],
|
| 54 |
+
output_dir=output,
|
| 55 |
+
crop_size=128,
|
| 56 |
+
limit=1,
|
| 57 |
+
)
|
| 58 |
+
|
| 59 |
+
print(datalist[0]["image"], datalist[0]["label"])
|
| 60 |
+
"""
|
| 61 |
+
|
| 62 |
+
if not len(datalist):
|
| 63 |
+
raise ValueError("Input datalist is empty")
|
| 64 |
+
|
| 65 |
+
if not loadmat:
|
| 66 |
+
print("Please make sure scipy with loadmat function is correctly installed")
|
| 67 |
+
raise ValueError("scipy.io.loadmat module/function not found")
|
| 68 |
+
|
| 69 |
+
dataset_json: List[Dict] = []
|
| 70 |
+
for d in tqdm(datalist):
|
| 71 |
+
logging.debug(f"Processing Image: {d['image']} => Label: {d['label']}")
|
| 72 |
+
|
| 73 |
+
# Image
|
| 74 |
+
image = PILImage.open(d["image"]).convert("RGB")
|
| 75 |
+
|
| 76 |
+
# Label
|
| 77 |
+
m = loadmat(d["label"])
|
| 78 |
+
instances = m["inst_map"]
|
| 79 |
+
|
| 80 |
+
for nuclei_id, (class_id, (y, x)) in enumerate(zip(m["inst_type"], m["inst_centroid"]), start=1):
|
| 81 |
+
x, y = (int(x), int(y))
|
| 82 |
+
class_id = int(class_id)
|
| 83 |
+
class_id = 3 if class_id in (3, 4) else 4 if class_id in (5, 6, 7) else class_id # override
|
| 84 |
+
|
| 85 |
+
if 0 < limit <= len(dataset_json):
|
| 86 |
+
return dataset_json
|
| 87 |
+
|
| 88 |
+
item = __prepare_patch(
|
| 89 |
+
d=d,
|
| 90 |
+
nuclei_id=nuclei_id,
|
| 91 |
+
output_dir=output_dir,
|
| 92 |
+
image=image,
|
| 93 |
+
instances=instances,
|
| 94 |
+
instance_idx=nuclei_id,
|
| 95 |
+
crop_size=crop_size,
|
| 96 |
+
class_id=class_id,
|
| 97 |
+
centroid=(x, y),
|
| 98 |
+
min_area=min_area,
|
| 99 |
+
min_distance=min_distance,
|
| 100 |
+
others_idx=255,
|
| 101 |
+
)
|
| 102 |
+
|
| 103 |
+
if item:
|
| 104 |
+
dataset_json.append(item)
|
| 105 |
+
|
| 106 |
+
return dataset_json
|
| 107 |
+
|
| 108 |
+
|
| 109 |
+
def __prepare_patch(
|
| 110 |
+
d,
|
| 111 |
+
nuclei_id,
|
| 112 |
+
output_dir,
|
| 113 |
+
image,
|
| 114 |
+
instances,
|
| 115 |
+
instance_idx,
|
| 116 |
+
crop_size,
|
| 117 |
+
class_id,
|
| 118 |
+
centroid,
|
| 119 |
+
min_area,
|
| 120 |
+
min_distance,
|
| 121 |
+
others_idx=255,
|
| 122 |
+
):
|
| 123 |
+
image_np = np.array(image)
|
| 124 |
+
image_size = image.size
|
| 125 |
+
|
| 126 |
+
bbox = __compute_bbox(crop_size, centroid, image_size)
|
| 127 |
+
|
| 128 |
+
cropped_label_np = instances[bbox[0] : bbox[2], bbox[1] : bbox[3]]
|
| 129 |
+
cropped_label_np = np.array(cropped_label_np)
|
| 130 |
+
|
| 131 |
+
this_label = np.where(cropped_label_np == instance_idx, class_id, 0)
|
| 132 |
+
if np.count_nonzero(this_label) < min_area:
|
| 133 |
+
return None
|
| 134 |
+
|
| 135 |
+
x, y = centroid
|
| 136 |
+
if x < min_distance or y < min_distance or (image_size[0] - x) < min_distance or (image_size[1] - y < min_distance):
|
| 137 |
+
return None
|
| 138 |
+
|
| 139 |
+
centroid = centroid[0] - bbox[0], centroid[1] - bbox[1]
|
| 140 |
+
others = np.where(np.logical_and(cropped_label_np > 0, cropped_label_np != instance_idx), others_idx, 0)
|
| 141 |
+
cropped_label_np = this_label + others
|
| 142 |
+
cropped_label = PILImage.fromarray(cropped_label_np.astype(np.uint8), None)
|
| 143 |
+
|
| 144 |
+
cropped_image_np = image_np[bbox[0] : bbox[2], bbox[1] : bbox[3], :]
|
| 145 |
+
cropped_image = PILImage.fromarray(cropped_image_np, "RGB")
|
| 146 |
+
|
| 147 |
+
images_dir = os.path.join(output_dir, "Images") if output_dir else "Images"
|
| 148 |
+
labels_dir = os.path.join(output_dir, "Labels") if output_dir else "Labels"
|
| 149 |
+
centroids_dir = os.path.join(output_dir, "Centroids") if output_dir else "Centroids"
|
| 150 |
+
|
| 151 |
+
os.makedirs(images_dir, exist_ok=True)
|
| 152 |
+
os.makedirs(labels_dir, exist_ok=True)
|
| 153 |
+
os.makedirs(centroids_dir, exist_ok=True)
|
| 154 |
+
|
| 155 |
+
image_id = pathlib.Path(d["image"]).stem
|
| 156 |
+
file_prefix = f"{image_id}_{class_id}_{str(instance_idx).zfill(4)}"
|
| 157 |
+
image_file = os.path.join(images_dir, f"{file_prefix}.png")
|
| 158 |
+
label_file = os.path.join(labels_dir, f"{file_prefix}.png")
|
| 159 |
+
centroid_file = os.path.join(centroids_dir, f"{file_prefix}.txt")
|
| 160 |
+
|
| 161 |
+
cropped_image.save(image_file)
|
| 162 |
+
cropped_label.save(label_file)
|
| 163 |
+
with open(centroid_file, "w") as fp:
|
| 164 |
+
json.dump([centroid], fp)
|
| 165 |
+
|
| 166 |
+
item = copy.deepcopy(d)
|
| 167 |
+
item["nuclei_id"] = nuclei_id
|
| 168 |
+
item["mask_value"] = class_id
|
| 169 |
+
item["image"] = image_file
|
| 170 |
+
item["label"] = label_file
|
| 171 |
+
item["centroid"] = centroid
|
| 172 |
+
return item
|
| 173 |
+
|
| 174 |
+
|
| 175 |
+
def __compute_bbox(patch_size, centroid, size):
|
| 176 |
+
x, y = centroid
|
| 177 |
+
m, n = size
|
| 178 |
+
|
| 179 |
+
x_start = int(max(x - patch_size / 2, 0))
|
| 180 |
+
y_start = int(max(y - patch_size / 2, 0))
|
| 181 |
+
x_end = x_start + patch_size
|
| 182 |
+
y_end = y_start + patch_size
|
| 183 |
+
if x_end > m:
|
| 184 |
+
x_end = m
|
| 185 |
+
x_start = m - patch_size
|
| 186 |
+
if y_end > n:
|
| 187 |
+
y_end = n
|
| 188 |
+
y_start = n - patch_size
|
| 189 |
+
return x_start, y_start, x_end, y_end
|
scripts/handlers.py
ADDED
|
@@ -0,0 +1,141 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
# Copyright (c) MONAI Consortium
|
| 2 |
+
# Licensed under the Apache License, Version 2.0 (the "License");
|
| 3 |
+
# you may not use this file except in compliance with the License.
|
| 4 |
+
# You may obtain a copy of the License at
|
| 5 |
+
# http://www.apache.org/licenses/LICENSE-2.0
|
| 6 |
+
# Unless required by applicable law or agreed to in writing, software
|
| 7 |
+
# distributed under the License is distributed on an "AS IS" BASIS,
|
| 8 |
+
# WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.
|
| 9 |
+
# See the License for the specific language governing permissions and
|
| 10 |
+
# limitations under the License.
|
| 11 |
+
|
| 12 |
+
import logging
|
| 13 |
+
from typing import TYPE_CHECKING, Any, Callable, List, Optional
|
| 14 |
+
|
| 15 |
+
import numpy as np
|
| 16 |
+
import torch
|
| 17 |
+
import torch.distributed
|
| 18 |
+
from monai.utils import IgniteInfo, min_version, optional_import
|
| 19 |
+
from sklearn.metrics import classification_report
|
| 20 |
+
|
| 21 |
+
Events, _ = optional_import("ignite.engine", IgniteInfo.OPT_IMPORT_VERSION, min_version, "Events")
|
| 22 |
+
make_grid, _ = optional_import("torchvision.utils", name="make_grid")
|
| 23 |
+
Image, _ = optional_import("PIL.Image")
|
| 24 |
+
ImageDraw, _ = optional_import("PIL.ImageDraw")
|
| 25 |
+
|
| 26 |
+
if TYPE_CHECKING:
|
| 27 |
+
from ignite.engine import Engine
|
| 28 |
+
from torch.utils.tensorboard import SummaryWriter
|
| 29 |
+
else:
|
| 30 |
+
Engine, _ = optional_import("ignite.engine", IgniteInfo.OPT_IMPORT_VERSION, min_version, "Engine")
|
| 31 |
+
SummaryWriter, _ = optional_import("torch.utils.tensorboard", name="SummaryWriter")
|
| 32 |
+
|
| 33 |
+
|
| 34 |
+
class TensorBoardImageHandler:
|
| 35 |
+
def __init__(
|
| 36 |
+
self,
|
| 37 |
+
class_names,
|
| 38 |
+
summary_writer: Optional[SummaryWriter] = None,
|
| 39 |
+
log_dir: str = "./runs",
|
| 40 |
+
tag_name="val",
|
| 41 |
+
interval: int = 1,
|
| 42 |
+
batch_transform: Callable = lambda x: x,
|
| 43 |
+
output_transform: Callable = lambda x: x,
|
| 44 |
+
batch_limit=1,
|
| 45 |
+
device=None,
|
| 46 |
+
) -> None:
|
| 47 |
+
self.class_names = class_names
|
| 48 |
+
self.writer = SummaryWriter(log_dir=log_dir) if summary_writer is None else summary_writer
|
| 49 |
+
self.tag_name = tag_name
|
| 50 |
+
self.interval = interval
|
| 51 |
+
self.batch_transform = batch_transform
|
| 52 |
+
self.output_transform = output_transform
|
| 53 |
+
self.batch_limit = batch_limit
|
| 54 |
+
self.device = device
|
| 55 |
+
|
| 56 |
+
self.logger = logging.getLogger(__name__)
|
| 57 |
+
|
| 58 |
+
if torch.distributed.is_initialized():
|
| 59 |
+
self.tag_name = f"{self.tag_name}-r{torch.distributed.get_rank()}"
|
| 60 |
+
self.class_y: List[Any] = []
|
| 61 |
+
self.class_y_pred: List[Any] = []
|
| 62 |
+
|
| 63 |
+
def attach(self, engine: Engine) -> None:
|
| 64 |
+
if self.interval == 1:
|
| 65 |
+
engine.add_event_handler(Events.ITERATION_COMPLETED(every=self.interval), self, "iteration")
|
| 66 |
+
engine.add_event_handler(Events.EPOCH_COMPLETED(every=self.interval), self, "epoch")
|
| 67 |
+
|
| 68 |
+
def __call__(self, engine: Engine, action) -> None:
|
| 69 |
+
epoch = engine.state.epoch
|
| 70 |
+
batch_data = self.batch_transform(engine.state.batch)
|
| 71 |
+
output_data = self.output_transform(engine.state.output)
|
| 72 |
+
|
| 73 |
+
if action == "iteration":
|
| 74 |
+
for bidx in range(len(batch_data)):
|
| 75 |
+
y = output_data[bidx]["label"].detach().cpu().numpy()
|
| 76 |
+
y_pred = output_data[bidx]["pred"].detach().cpu().numpy()
|
| 77 |
+
|
| 78 |
+
self.class_y.append(np.argmax(y))
|
| 79 |
+
self.class_y_pred.append(np.argmax(y_pred))
|
| 80 |
+
return
|
| 81 |
+
|
| 82 |
+
self.write_metrics(epoch)
|
| 83 |
+
self.write_images(batch_data, output_data, epoch)
|
| 84 |
+
self.writer.flush()
|
| 85 |
+
|
| 86 |
+
def write_images(self, batch_data, output_data, epoch):
|
| 87 |
+
for bidx in range(len(batch_data)):
|
| 88 |
+
image = batch_data[bidx]["image"].detach().cpu().numpy()
|
| 89 |
+
y = output_data[bidx]["label"].detach().cpu().numpy()
|
| 90 |
+
y_pred = output_data[bidx]["pred"].detach().cpu().numpy()
|
| 91 |
+
|
| 92 |
+
sig_np = image[:3] * 128 + 128
|
| 93 |
+
sig_np[0, :, :] = np.where(image[3] > 0, 1, sig_np[0, :, :])
|
| 94 |
+
|
| 95 |
+
y_c = np.argmax(y)
|
| 96 |
+
y_pred_c = np.argmax(y_pred)
|
| 97 |
+
|
| 98 |
+
tag_prefix = f"{self.tag_name} - b{bidx} - " if self.batch_limit != 1 else f"{self.tag_name} - "
|
| 99 |
+
label_pred_tag = f"{tag_prefix}Image/Signal/Label/Pred:"
|
| 100 |
+
|
| 101 |
+
y_img = Image.new("RGB", image.shape[-2:])
|
| 102 |
+
draw = ImageDraw.Draw(y_img)
|
| 103 |
+
draw.text((10, 50), self.class_names.get(f"{y_c}", f"{y_c}"))
|
| 104 |
+
|
| 105 |
+
y_pred_img = Image.new("RGB", image.shape[-2:], "green" if y_c == y_pred_c else "red")
|
| 106 |
+
draw = ImageDraw.Draw(y_pred_img)
|
| 107 |
+
draw.text((10, 50), self.class_names.get(f"{y_pred_c}", f"{y_pred_c}"))
|
| 108 |
+
|
| 109 |
+
img_tensor = make_grid(
|
| 110 |
+
tensor=[
|
| 111 |
+
torch.from_numpy(sig_np),
|
| 112 |
+
torch.from_numpy(np.stack((np.where(image[3] > 0, 255, 0),) * 3)),
|
| 113 |
+
torch.from_numpy(np.moveaxis(np.array(y_img), -1, 0)),
|
| 114 |
+
torch.from_numpy(np.moveaxis(np.array(y_pred_img), -1, 0)),
|
| 115 |
+
],
|
| 116 |
+
nrow=4,
|
| 117 |
+
normalize=True,
|
| 118 |
+
pad_value=10,
|
| 119 |
+
)
|
| 120 |
+
self.writer.add_image(tag=label_pred_tag, img_tensor=img_tensor, global_step=epoch)
|
| 121 |
+
|
| 122 |
+
if self.batch_limit == 1 or bidx == (self.batch_limit - 1):
|
| 123 |
+
break
|
| 124 |
+
|
| 125 |
+
def write_metrics(self, epoch):
|
| 126 |
+
cr = classification_report(self.class_y, self.class_y_pred, output_dict=True, zero_division=0)
|
| 127 |
+
for k, v in cr.items():
|
| 128 |
+
if isinstance(v, dict):
|
| 129 |
+
ltext = []
|
| 130 |
+
cname = self.class_names.get(k, k)
|
| 131 |
+
for n, m in v.items():
|
| 132 |
+
ltext.append(f"{n} => {m:.4f}")
|
| 133 |
+
self.writer.add_scalar(f"{self.tag_name}_cr_{cname}_{n}", m, epoch)
|
| 134 |
+
|
| 135 |
+
self.logger.info(f"{self.tag_name} => Epoch[{epoch}] - {cname} - Metrics -- {'; '.join(ltext)}")
|
| 136 |
+
else:
|
| 137 |
+
self.logger.info(f"{self.tag_name} => Epoch[{epoch}] Metrics -- {k} => {v:.4f}")
|
| 138 |
+
self.writer.add_scalar(f"{self.tag_name}_cr_{k}", v, epoch)
|
| 139 |
+
|
| 140 |
+
self.class_y = []
|
| 141 |
+
self.class_y_pred = []
|
scripts/writer.py
ADDED
|
@@ -0,0 +1,54 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
# Copyright (c) MONAI Consortium
|
| 2 |
+
# Licensed under the Apache License, Version 2.0 (the "License");
|
| 3 |
+
# you may not use this file except in compliance with the License.
|
| 4 |
+
# You may obtain a copy of the License at
|
| 5 |
+
# http://www.apache.org/licenses/LICENSE-2.0
|
| 6 |
+
# Unless required by applicable law or agreed to in writing, software
|
| 7 |
+
# distributed under the License is distributed on an "AS IS" BASIS,
|
| 8 |
+
# WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.
|
| 9 |
+
# See the License for the specific language governing permissions and
|
| 10 |
+
# limitations under the License.
|
| 11 |
+
import json
|
| 12 |
+
import logging
|
| 13 |
+
from typing import Dict, Mapping, Optional
|
| 14 |
+
|
| 15 |
+
import numpy as np
|
| 16 |
+
from monai.config import NdarrayOrTensor, PathLike
|
| 17 |
+
from monai.data import ImageWriter
|
| 18 |
+
|
| 19 |
+
logger = logging.getLogger(__name__)
|
| 20 |
+
|
| 21 |
+
|
| 22 |
+
class ClassificationWriter(ImageWriter):
|
| 23 |
+
def __init__(self, label_index_map: Optional[Dict[str, str]] = None, **kwargs):
|
| 24 |
+
super().__init__(**kwargs)
|
| 25 |
+
self.label_index_map = (
|
| 26 |
+
label_index_map
|
| 27 |
+
if label_index_map
|
| 28 |
+
else {"0": "Other", "1": "Inflammatory", "2": "Epithelial", "3": "Spindle-Shaped"}
|
| 29 |
+
)
|
| 30 |
+
|
| 31 |
+
def set_data_array(
|
| 32 |
+
self,
|
| 33 |
+
data_array: NdarrayOrTensor,
|
| 34 |
+
channel_dim: Optional[int] = 0,
|
| 35 |
+
squeeze_end_dims: bool = True,
|
| 36 |
+
contiguous: bool = False,
|
| 37 |
+
**kwargs,
|
| 38 |
+
):
|
| 39 |
+
self.data_obj: np.ndarray = super().create_backend_obj(data_array)
|
| 40 |
+
|
| 41 |
+
def set_metadata(self, meta_dict: Optional[Mapping] = None, resample: bool = True, **options):
|
| 42 |
+
pass
|
| 43 |
+
|
| 44 |
+
def write(self, filename: PathLike, verbose: bool = False, **kwargs):
|
| 45 |
+
super().write(filename, verbose=verbose)
|
| 46 |
+
result = []
|
| 47 |
+
for idx, score in enumerate(self.data_obj):
|
| 48 |
+
name = f"label_{idx}"
|
| 49 |
+
name = self.label_index_map.get(str(idx)) if self.label_index_map else name
|
| 50 |
+
if name:
|
| 51 |
+
result.append({"idx": idx, "label": name, "score": float(score)})
|
| 52 |
+
|
| 53 |
+
with open(filename, "w") as fp:
|
| 54 |
+
json.dump(result, fp)
|